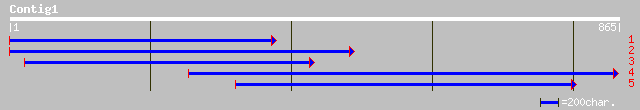
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001587A_C01 KCC001587A_c01
(805 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
dbj|BAB90282.1| P0470A12.7 [Oryza sativa (japonica cultivar-group)] 67 3e-10
ref|XP_308231.1| ENSANGP00000020783 [Anopheles gambiae] gi|21291... 65 2e-09
ref|NP_491624.1| ribophorin II (31.1 kD) (1G118) [Caenorhabditis... 59 7e-08
gb|AAH46727.1| Similar to ribophorin II [Xenopus laevis] 59 7e-08
ref|NP_193847.2| expressed protein [Arabidopsis thaliana] gi|166... 57 3e-07
>dbj|BAB90282.1| P0470A12.7 [Oryza sativa (japonica cultivar-group)]
Length = 689
Score = 67.4 bits (163), Expect = 3e-10
Identities = 46/137 (33%), Positives = 68/137 (49%), Gaps = 3/137 (2%)
Frame = +1
Query: 382 GESFKPQQVMLMLKSKSLGLAAYAVGKLKSGSYSLSINAAAVEKQIGKAAGEYEATLLVG 561
G++FKP QV L LK + V + + + ++ + ++ +G Y+ L VG
Sbjct: 472 GKTFKPHQVFLKLKHDESKVEHLFVVPGSARQFKIVLDFLGLVEKFYYLSGRYDLELAVG 531
Query: 562 DPSTPKGISYALGTAEL-LFSGASNAPEPAAAVRT--AKFQPANNVKPDITHIFRAPEKW 732
D + ALG EL L AP+P A +KF P K +I+HIFR+PEK
Sbjct: 532 DAAMENSFLRALGHIELDLPEAPEKAPKPPAQAVDPFSKFGP----KKEISHIFRSPEKR 587
Query: 733 PPAIVSLVFAGLALVPL 783
PP +S F GL L+P+
Sbjct: 588 PPKELSFAFTGLTLLPI 604
>ref|XP_308231.1| ENSANGP00000020783 [Anopheles gambiae] gi|21291954|gb|EAA04099.1|
ENSANGP00000020783 [Anopheles gambiae str. PEST]
Length = 671
Score = 64.7 bits (156), Expect = 2e-09
Identities = 50/199 (25%), Positives = 84/199 (42%), Gaps = 5/199 (2%)
Frame = +1
Query: 211 QAQVVAELSISDVKVVIKGADGSAISTLTGT--YPK--AVLANPQVPAAGSLDVSLSVKK 378
Q V+ ++ +S ++V + D +A S + +P A + N L +L
Sbjct: 401 QVSVLGKVRVSSLEVGVGDTDSAASSVKRQSVAHPSKLATVLNADSQQKVVLKTALVDDA 460
Query: 379 GGESFKPQQVMLMLKSKSLGLAAYAVGKLKSG-SYSLSINAAAVEKQIGKAAGEYEATLL 555
G+ Q ++L+ + V + S +Y ++ A G +G YE L+
Sbjct: 461 SGKPITVHQCFVLLRHQETRQEIIFVAETDSSKAYKFEMDVGARAADFGHKSGLYEVRLI 520
Query: 556 VGDPSTPKGISYALGTAELLFSGASNAPEPAAAVRTAKFQPANNVKPDITHIFRAPEKWP 735
VGD + L EL F+ + + + + +P P+I H FRAPEK P
Sbjct: 521 VGDALLSNSFQWHLADLELKFAAGDSQQQQSVDAAASSRKPL----PEIVHQFRAPEKRP 576
Query: 736 PAIVSLVFAGLALVPLAIV 792
P VS +F L + PL I+
Sbjct: 577 PRFVSDLFTALCIAPLVIL 595
>ref|NP_491624.1| ribophorin II (31.1 kD) (1G118) [Caenorhabditis elegans]
gi|7505848|pir||T25829 hypothetical protein M01A10.3 -
Caenorhabditis elegans gi|1825655|gb|AAB42275.1|
Hypothetical protein M01A10.3 [Caenorhabditis elegans]
Length = 280
Score = 59.3 bits (142), Expect = 7e-08
Identities = 44/155 (28%), Positives = 74/155 (47%), Gaps = 2/155 (1%)
Frame = +1
Query: 340 AAGSLDVSLSVKKGGES--FKPQQVMLMLKSKSLGLAAYAVGKLKSGSYSLSINAAAVEK 513
A+ L VS +V K ++ KP QV L +++ V +G+Y K
Sbjct: 59 ASQRLYVSFTVAKKSDNAKVKPHQVFLRFVAQNGEDVVVVVNPDANGNYVYDNVLRTAAK 118
Query: 514 QIGKAAGEYEATLLVGDPSTPKGISYALGTAELLFSGASNAPEPAAAVRTAKFQPANNVK 693
+G+++ +LLVGD + I++ + + A EP + F+P N
Sbjct: 119 SFRNLSGQFKISLLVGDVTIKNPINWQFANID---AALPVAYEPTPKSQQVHFEPLN--- 172
Query: 694 PDITHIFRAPEKWPPAIVSLVFAGLALVPLAIVIL 798
+I+HIFR PEK P A++S +F + L PL I+++
Sbjct: 173 -EISHIFRQPEKRPSALISDLFTIICLSPLLILVV 206
>gb|AAH46727.1| Similar to ribophorin II [Xenopus laevis]
Length = 631
Score = 59.3 bits (142), Expect = 7e-08
Identities = 53/206 (25%), Positives = 87/206 (41%), Gaps = 11/206 (5%)
Frame = +1
Query: 214 AQVVAELSISDVKVVIKGADGSAISTLTGTYPKAVLANPQVPAAG----------SLDVS 363
+QV ++ +S +V I D S + PK N A G +L
Sbjct: 361 SQVELKVKVS-TEVGIGNVDLSIVDKDQSISPKMTRVNFPSKAKGPFTTDSHQNFALSFQ 419
Query: 364 LSVKKGGESFKPQQVMLMLKSKSLGLAAYAVGKLKS-GSYSLSINAAAVEKQIGKAAGEY 540
L+ G S P Q + L ++ G V + + +Y ++ + + + A+G Y
Sbjct: 420 LTDVNTGASLTPHQTFVRLHNQKTGQEVVFVAEPDNKNTYRFELDPSERKSEFDSASGTY 479
Query: 541 EATLLVGDPSTPKGISYALGTAELLFSGASNAPEPAAAVRTAKFQPANNVKPDITHIFRA 720
L+VGD + + + + + F ++P P + F P KPDI H+FR
Sbjct: 480 TLYLIVGDATIENPMLWNVADVLIKFPD-EDSPVPIQSKNL--FIP----KPDIQHLFRE 532
Query: 721 PEKWPPAIVSLVFAGLALVPLAIVIL 798
PEK PP +VS F L L PL ++ +
Sbjct: 533 PEKRPPTVVSNTFTALILAPLLLLFI 558
>ref|NP_193847.2| expressed protein [Arabidopsis thaliana] gi|16604454|gb|AAL24233.1|
AT4g21150/F7J7_90 [Arabidopsis thaliana]
gi|18958022|gb|AAL79584.1| AT4g21150/F7J7_90
[Arabidopsis thaliana]
Length = 691
Score = 57.4 bits (137), Expect = 3e-07
Identities = 41/135 (30%), Positives = 63/135 (46%), Gaps = 2/135 (1%)
Frame = +1
Query: 382 GESFKPQQVMLMLKSKSLGLAAYAVGKLKSGSYSLSINAAAVEKQIGKAAGEYEATLLVG 561
G +FKP Q LK +S + V K L ++ + +++ +G+YE L +G
Sbjct: 475 GNAFKPHQAFFKLKHESQVEHIFLV-KTSGKKSELVLDFLGLVEKLYYLSGKYEIQLTIG 533
Query: 562 DPSTPKGISYALGTAELLFSGASNAPEPAAAVRTAKFQPANNVKP--DITHIFRAPEKWP 735
D S + +G EL PE A +P + P +I+HIFR PEK P
Sbjct: 534 DASMENSLLSNIGHIEL---DLPERPEKATRPPLQSTEPYSRYGPKAEISHIFRIPEKLP 590
Query: 736 PAIVSLVFAGLALVP 780
+SLVF G+ ++P
Sbjct: 591 AKQLSLVFLGVIVLP 605