Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001563A_C01 KCC001563A_c01
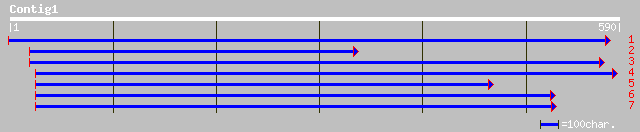
(590 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG25917.1|AF190427_1 putative GAL4-like transcriptional acti... 40 0.019
ref|ZP_00058502.1| hypothetical protein [Thermobifida fusca] 40 0.024
ref|XP_330349.1| hypothetical protein [Neurospora crassa] gi|289... 40 0.032
gb|AAK29456.1|AF352253_1 histone H1 [Lens culinaris] 39 0.041
gb|AAK29455.1|AF352252_1 histone H1 [Lens culinaris] 39 0.041
>gb|AAG25917.1|AF190427_1 putative GAL4-like transcriptional activator [Colletotrichum
lindemuthianum]
Length = 746
Score = 40.4 bits (93), Expect = 0.019
Identities = 30/104 (28%), Positives = 44/104 (41%), Gaps = 8/104 (7%)
Frame = +2
Query: 227 HTPQVHSKLNSSQT*ATALLQEPHLLRHSANVAPERPAHPAIRKSCRRIPPPRP--PHPH 400
H PQ H + Q + H L+H P HP +S P P+P PHPH
Sbjct: 630 HQPQQHQQQQHQQ--------QQHQLQHQQFSPGPPPPHPHHHQSHASYPTPQPPQPHPH 681
Query: 401 HGPF--WPALRRSRHAASH---STPA-RLPYLPPPRARPITRWR 514
H P +P + H S PA ++P + P +P+ ++R
Sbjct: 682 HQPHPSYPPPQHQPHPHSQVIMGHPAMQIPGMMPQPGQPMVQYR 725
>ref|ZP_00058502.1| hypothetical protein [Thermobifida fusca]
Length = 1903
Score = 40.0 bits (92), Expect = 0.024
Identities = 38/125 (30%), Positives = 43/125 (34%), Gaps = 8/125 (6%)
Frame = +2
Query: 200 PTHT*KLLTHTPQVHSKLNSSQT*ATALLQEPHLLRHSANVAPERPAHPAIRKSCRR--- 370
P H + P H + A PH R + P P P R RR
Sbjct: 275 PRHARQTRVARPHHHRAQRRPRPRAAPAHPAPHRPRLPRDRLPRPPRRPETRHPRRRRGR 334
Query: 371 ---IPPPRPPHPHHGPFWPALRRSRHAASHSTPARLPYLPPPRARPITRW--RSGHGSRA 535
+P PRP PH G R R AA P R PPR RP+ R RSG
Sbjct: 335 RTPVPLPRPARPHRG----TRPRKRAAAGTHRPPR-----PPRRRPLARLPARSGKERGP 385
Query: 536 RGGAP 550
R P
Sbjct: 386 RRSHP 390
>ref|XP_330349.1| hypothetical protein [Neurospora crassa] gi|28920331|gb|EAA29702.1|
hypothetical protein [Neurospora crassa]
Length = 1278
Score = 39.7 bits (91), Expect = 0.032
Identities = 38/137 (27%), Positives = 51/137 (36%), Gaps = 3/137 (2%)
Frame = +2
Query: 89 RTKSKLFLQGGNPAPACPYKASHMFFLLASTHVST*SPTHT*KLLTH-TPQVHSKLNSSQ 265
RT S L P+ PY AS S T + H +LT +P ++
Sbjct: 414 RTYSIPALPPEPPSHIDPYAASSSHHYNQSHPHFTDASGHQISILTSPSPGPPMQVTPDT 473
Query: 266 T*ATALLQEPHLLRHSANVAPERPAHPAIRKSCRR-IPPPRPPHPHHGP-FWPALRRSRH 439
T + Q P A+ +P P HP+ PPP P HPHH P + S H
Sbjct: 474 TNLATVQQNPFTRHTQAHESPMLPHHPSSPPHHHHHYPPPYPSHPHHEPQHHSVVTSSHH 533
Query: 440 AASHSTPARLPYLPPPR 490
H LP + P+
Sbjct: 534 HQPHHPHVSLPPIIDPQ 550
>gb|AAK29456.1|AF352253_1 histone H1 [Lens culinaris]
Length = 293
Score = 39.3 bits (90), Expect = 0.041
Identities = 26/84 (30%), Positives = 39/84 (45%), Gaps = 1/84 (1%)
Frame = +1
Query: 280 PSARATPPAALRQCGT*APSAPCHP*ELPSHTATTTSTPTPWTILARPPPLQTRCIP-QH 456
P+A+A P A + P+A P P+ A T++ +P T A P P + P +
Sbjct: 210 PAAKAKPAAKAKPAAKAKPAAKAKPAAKPAKAARTSTRTSPGTRAAAPKPAAKKAAPAKK 269
Query: 457 ARTLAISSTATSTADYAVAFRSRQ 528
A A + TA S A A A R ++
Sbjct: 270 APVKAAAKTAKSPAKKAAAKRGKK 293
>gb|AAK29455.1|AF352252_1 histone H1 [Lens culinaris]
Length = 293
Score = 39.3 bits (90), Expect = 0.041
Identities = 26/84 (30%), Positives = 39/84 (45%), Gaps = 1/84 (1%)
Frame = +1
Query: 280 PSARATPPAALRQCGT*APSAPCHP*ELPSHTATTTSTPTPWTILARPPPLQTRCIP-QH 456
P+A+A P A + P+A P P+ A T++ +P T A P P + P +
Sbjct: 210 PAAKAKPAAKAKPAAKAKPAAKAKPAAKPAKAARTSTRTSPGTRAAAPKPAAKKAAPAKK 269
Query: 457 ARTLAISSTATSTADYAVAFRSRQ 528
A A + TA S A A A R ++
Sbjct: 270 APVKAAAKTAKSPAKKAAAKRGKK 293