Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001548A_C01 KCC001548A_c01
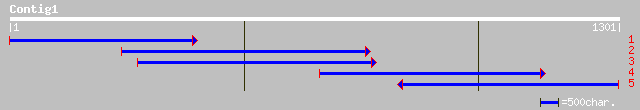
(1301 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAO20101.1| putative phosphate/phosphoenolpyruvate translocat... 313 4e-84
dbj|BAC75429.1| putative phosphoenolpyruvate/phosphate transloca... 169 8e-41
pir||T04096 glucose-6-phosphate/phosphate-translocator precursor... 168 2e-40
pir||T04100 glucose-6-phosphate/phosphate-translocator precursor... 167 3e-40
gb|AAK51561.1|AF372833_1 phosphoenolpyruvate/phosphate transloca... 167 4e-40
>gb|AAO20101.1| putative phosphate/phosphoenolpyruvate translocator precursor
protein [Chlamydomonas reinhardtii]
Length = 399
Score = 313 bits (802), Expect = 4e-84
Identities = 163/163 (100%), Positives = 163/163 (100%)
Frame = +3
Query: 3 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 182
LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV
Sbjct: 237 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 296
Query: 183 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 362
GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL
Sbjct: 297 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 356
Query: 363 FFRNPVSLQNALGTALALAGVFLYGTVKRQQAIAAGKKIAASE 491
FFRNPVSLQNALGTALALAGVFLYGTVKRQQAIAAGKKIAASE
Sbjct: 357 FFRNPVSLQNALGTALALAGVFLYGTVKRQQAIAAGKKIAASE 399
>dbj|BAC75429.1| putative phosphoenolpyruvate/phosphate translocator [Oryza sativa
(japonica cultivar-group)]
Length = 407
Score = 169 bits (429), Expect = 8e-41
Identities = 92/149 (61%), Positives = 110/149 (73%)
Frame = +3
Query: 3 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 182
+TFQSRNVLSKKLM+KK++ LDN+ LFS+IT++S LL P L EG K++P
Sbjct: 259 VTFQSRNVLSKKLMVKKEES-------LDNINLFSIITVMSFFLLAPVAFLTEGIKITPT 311
Query: 183 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 362
L G+ VL + +A LCFH YQQVSYMIL+RVSPVTHS+GNCVKRVVVI SVL
Sbjct: 312 VLQSAGLNVKQ-VLTRSLLAALCFHAYQQVSYMILARVSPVTHSVGNCVKRVVVIVTSVL 370
Query: 363 FFRNPVSLQNALGTALALAGVFLYGTVKR 449
FFR PVS N+LGTA+ALAGVFLY +KR
Sbjct: 371 FFRTPVSPINSLGTAIALAGVFLYSQLKR 399
>pir||T04096 glucose-6-phosphate/phosphate-translocator precursor homolog -
maize gi|1778147|gb|AAB40649.1|
phosphate/phosphoenolpyruvate translocator precursor
Length = 390
Score = 168 bits (425), Expect = 2e-40
Identities = 92/149 (61%), Positives = 109/149 (72%)
Frame = +3
Query: 3 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 182
+TFQSRNVLSKKLM+KK++ LDN+ LFS+IT++S LL P TLL EG K+SP
Sbjct: 242 VTFQSRNVLSKKLMVKKEES-------LDNINLFSIITVMSFFLLAPVTLLTEGVKVSPA 294
Query: 183 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 362
L G+ V + +A CFH YQQVSYMIL+RVSPVTHS+GNCVKRVVVI SVL
Sbjct: 295 VLQSAGLNLKQ-VYTRSLIAACCFHAYQQVSYMILARVSPVTHSVGNCVKRVVVIVTSVL 353
Query: 363 FFRNPVSLQNALGTALALAGVFLYGTVKR 449
FFR PVS N+LGT +ALAGVFLY +KR
Sbjct: 354 FFRTPVSPINSLGTGIALAGVFLYSQLKR 382
>pir||T04100 glucose-6-phosphate/phosphate-translocator precursor homolog -
maize gi|1778149|gb|AAB40650.1|
phosphate/phosphoenolpyruvate translocator precursor
Length = 396
Score = 167 bits (424), Expect = 3e-40
Identities = 91/149 (61%), Positives = 109/149 (73%)
Frame = +3
Query: 3 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 182
+TFQSRNVLSKKLM+KK++ LDN+ LFS+IT++S LL P TLL EG K+SP
Sbjct: 248 VTFQSRNVLSKKLMVKKEES-------LDNINLFSIITVMSFFLLAPVTLLTEGVKVSPA 300
Query: 183 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 362
L G+ + + +A CFH YQQVSYMIL+RVSPVTHS+GNCVKRVVVI SVL
Sbjct: 301 VLQSAGLNLKQ-IYTRSLIAACCFHAYQQVSYMILARVSPVTHSVGNCVKRVVVIVTSVL 359
Query: 363 FFRNPVSLQNALGTALALAGVFLYGTVKR 449
FFR PVS N+LGT +ALAGVFLY +KR
Sbjct: 360 FFRTPVSPINSLGTGIALAGVFLYSQLKR 388
>gb|AAK51561.1|AF372833_1 phosphoenolpyruvate/phosphate translocator [Oryza sativa]
Length = 408
Score = 167 bits (423), Expect = 4e-40
Identities = 90/149 (60%), Positives = 109/149 (72%)
Frame = +3
Query: 3 LTFQSRNVLSKKLMLKKKDKDGNAEAPLDNMALFSVITLLSAALLLPATLLFEGWKLSPV 182
+TFQSRNVLSKKLM+KK++ LDN+ LFS+IT++S LL P TLL EG K++P
Sbjct: 260 VTFQSRNVLSKKLMVKKEES-------LDNITLFSIITVMSFFLLAPVTLLTEGVKVTPT 312
Query: 183 GLAEMGVRSPNGVLAHAAMAGLCFHLYQQVSYMILSRVSPVTHSIGNCVKRVVVIAASVL 362
L G+ + + +A CFH YQQVSYMIL+RVSPVTHS+GNCVKRVVVI SVL
Sbjct: 313 VLQSAGLNLKQ-IYTRSLIAAFCFHAYQQVSYMILARVSPVTHSVGNCVKRVVVIVTSVL 371
Query: 363 FFRNPVSLQNALGTALALAGVFLYGTVKR 449
FFR PVS N+LGT +ALAGVFLY +KR
Sbjct: 372 FFRTPVSPINSLGTGVALAGVFLYSQLKR 400