Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001406A_C01 KCC001406A_c01
(822 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
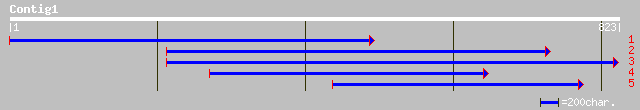
ref|XP_309353.1| ENSANGP00000015008 [Anopheles gambiae] gi|21292... 40 0.060
ref|ZP_00040388.1| COG3525: N-acetyl-beta-hexosaminidase [Xylell... 31 0.083
ref|NP_780011.1| beta-hexosaminidase precursor [Xylella fastidio... 31 0.11
ref|NP_298137.1| beta-hexosaminidase precursor [Xylella fastidio... 29 0.14
gb|AAA75509.1| orf 38 0.17
>ref|XP_309353.1| ENSANGP00000015008 [Anopheles gambiae] gi|21292910|gb|EAA05055.1|
ENSANGP00000015008 [Anopheles gambiae str. PEST]
Length = 372
Score = 39.7 bits (91), Expect = 0.060
Identities = 49/188 (26%), Positives = 70/188 (37%), Gaps = 11/188 (5%)
Frame = -1
Query: 618 PHLVPIVTRVIRPAGPATA---TTSRPSDTPT-----GGAAIRLAPAL---LDAHLAHTH 472
P + PI++ ++ P P T+ T S PS PT AA APA LD A T
Sbjct: 176 PMMPPIISYLLPPMPPTTSVPSTMSSPSAIPTTSTSSAAAAAHNAPASMPSLDTVYASTS 235
Query: 471 MHARFTPPTTLLRHTQDPRNNPAQRRRTVNLEPPTTFLAWHKFQVLVRRAGVQPGQRH*L 292
M + P+T R N P PPTT A G +
Sbjct: 236 MMPPPSTPSTSNRAAAS--NQPTS--------PPTT-------------AANTNGSKRGS 272
Query: 291 RASMATASACLSSHPPPTPPSRRYRQQIGNIVFFCCCAKRKPAFPSINPPCGSNSHPIPV 112
RA+ ++ + + PPTPP+ + R + ++ P S PP P P+
Sbjct: 273 RAAPKSSPSTSRTPEPPTPPAPKRRNRRASVAASQSPYLTAPPSQSPIPPLPPTPRPPPI 332
Query: 111 ASRVGANL 88
A+ L
Sbjct: 333 ATPTDTTL 340
>ref|ZP_00040388.1| COG3525: N-acetyl-beta-hexosaminidase [Xylella fastidiosa Ann-1]
Length = 812
Score = 31.2 bits (69), Expect(2) = 0.083
Identities = 18/59 (30%), Positives = 26/59 (43%)
Frame = -1
Query: 576 GPATATTSRPSDTPTGGAAIRLAPALLDAHLAHTHMHARFTPPTTLLRHTQDPRNNPAQ 400
GP T + P+DT +A LA L H+H TP +R +DP++ Q
Sbjct: 77 GPHTLISIPPNDTDAQHSATYLATLLQHTRNLTLHIHTETTPKPDSIRLQRDPQSPVTQ 135
Score = 26.9 bits (58), Expect(2) = 0.083
Identities = 22/65 (33%), Positives = 29/65 (43%)
Frame = -3
Query: 805 PGALLPLLMFRAMRKLCLLLYNAPAFPSHDSHEKNSWLRVKPIRA*RAPIQCRALGHVLP 626
P LPLL RA+ LC+ + +F + SH K S + P A P+ LP
Sbjct: 8 PTQALPLL--RALLTLCIAALSTTSFDTTASHHKKSQHPLPPRTASSPPLP------PLP 59
Query: 625 DNPAP 611
PAP
Sbjct: 60 LIPAP 64
>ref|NP_780011.1| beta-hexosaminidase precursor [Xylella fastidiosa Temecula1]
gi|28057818|gb|AAO29660.1| beta-hexosaminidase precursor
[Xylella fastidiosa Temecula1]
Length = 812
Score = 31.2 bits (69), Expect(2) = 0.11
Identities = 18/59 (30%), Positives = 26/59 (43%)
Frame = -1
Query: 576 GPATATTSRPSDTPTGGAAIRLAPALLDAHLAHTHMHARFTPPTTLLRHTQDPRNNPAQ 400
GP T + P+DT +A LA L H+H TP +R +DP++ Q
Sbjct: 77 GPHTLISIPPNDTDAQHSATYLATLLQHTRNLTLHIHTETTPTPDSIRLQRDPQSPVTQ 135
Score = 26.6 bits (57), Expect(2) = 0.11
Identities = 22/65 (33%), Positives = 29/65 (43%)
Frame = -3
Query: 805 PGALLPLLMFRAMRKLCLLLYNAPAFPSHDSHEKNSWLRVKPIRA*RAPIQCRALGHVLP 626
P LPLL RA+ LC+ + +F + SH K S + P A P+ LP
Sbjct: 8 PTQALPLL--RALLTLCIAALSTTSFDTTASHHKKSQHPLPPRTASGPPLP------PLP 59
Query: 625 DNPAP 611
PAP
Sbjct: 60 LIPAP 64
>ref|NP_298137.1| beta-hexosaminidase precursor [Xylella fastidiosa 9a5c]
gi|11360518|pir||B82755 beta-hexosaminidase precursor
XF0847 [imported] - Xylella fastidiosa (strain 9a5c)
gi|9105753|gb|AAF83657.1|AE003924_3 beta-hexosaminidase
precursor [Xylella fastidiosa 9a5c]
Length = 841
Score = 29.3 bits (64), Expect(2) = 0.14
Identities = 27/79 (34%), Positives = 34/79 (42%), Gaps = 6/79 (7%)
Frame = -3
Query: 808 RPGALLPLLMFRAMRKLCLLLYNAPAFPSHDSHEKNSWLRVKPIRA*RAPIQCRALGHVL 629
RP LPLL RA+ LC+ + +F + SH K S P+ AP G L
Sbjct: 39 RPTQALPLL--RALLTLCIAALSTTSFDTTASHHKKS---QHPLPPRTAP------GPPL 87
Query: 628 PDNPAP------RSHCHAG 590
P PAP + H H G
Sbjct: 88 PLIPAPVQIQRGQGHIHIG 106
Score = 28.1 bits (61), Expect(2) = 0.14
Identities = 17/59 (28%), Positives = 25/59 (41%)
Frame = -1
Query: 576 GPATATTSRPSDTPTGGAAIRLAPALLDAHLAHTHMHARFTPPTTLLRHTQDPRNNPAQ 400
G T + P+DT +A LA L H+H TP +R +DP++ Q
Sbjct: 106 GTDTLISIPPNDTDAQHSATYLATLLQHTRNLTLHIHTETTPTPDSIRLQRDPQSPVTQ 164
>gb|AAA75509.1| orf
Length = 404
Score = 38.1 bits (87), Expect = 0.17
Identities = 47/163 (28%), Positives = 58/163 (34%), Gaps = 11/163 (6%)
Frame = -1
Query: 696 GFVSSLSVHDVHPSNAELLGMCCRITPHLVPIVTRVIRP------AGPATATTSRPSDTP 535
G V L D P G+ + P +P + R RP A P S P+ P
Sbjct: 17 GRVPGLRSGDGRPRRVSRTGIAEDVDP--LPGLPRRARPSVSGYAARPGALAVSGPAGQP 74
Query: 534 TGGAAIRLAPALLDAHLAHTHMHARFT----PPTTLLRHTQDPRNNPAQRRRTVNLEPPT 367
G R DA A A T PP RH P N PA+R N P +
Sbjct: 75 PGQGPQRPRRPAKDAQPAPPPRPAGATRTGAPPAEGRRHKAAPHNGPARRSPPCNPAPRS 134
Query: 366 TFLAWHKFQVLVRRAGVQPGQRH*L-RASMATASACLSSHPPP 241
W R G + + L AS+ TAS+ L PPP
Sbjct: 135 CTHIW-----ATRACGPRAPRSSALGSASVPTASSRLRDGPPP 172