Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001386A_C01 KCC001386A_c01
(680 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
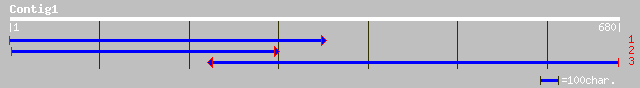
gb|AAK72125.1| outer dynein arm docking complex protein ODA1 [Ch... 135 4e-31
gb|AAH46717.1| Similar to zinc finger protein 162 [Xenopus laevis] 51 1e-05
ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca] 50 2e-05
emb|CAD70966.1| probable Cytokinesis protein sepA [Neurospora cr... 49 5e-05
ref|XP_327870.1| hypothetical protein [Neurospora crassa] gi|289... 49 5e-05
>gb|AAK72125.1| outer dynein arm docking complex protein ODA1 [Chlamydomonas
reinhardtii]
Length = 552
Score = 135 bits (341), Expect = 4e-31
Identities = 67/68 (98%), Positives = 67/68 (98%)
Frame = -2
Query: 679 IIEPPSTTQEEEVEGLEPEPVEEDRPLTREHLESKVQRTLPRKLETAIKVRPAGADATGG 500
IIEPPST QEEEVEGLEPEPVEEDRPLTREHLESKVQRTLPRKLETAIKVRPAGADATGG
Sbjct: 485 IIEPPSTMQEEEVEGLEPEPVEEDRPLTREHLESKVQRTLPRKLETAIKVRPAGADATGG 544
Query: 499 KRGSPTRR 476
KRGSPTRR
Sbjct: 545 KRGSPTRR 552
>gb|AAH46717.1| Similar to zinc finger protein 162 [Xenopus laevis]
Length = 571
Score = 51.2 bits (121), Expect = 1e-05
Identities = 45/135 (33%), Positives = 56/135 (41%), Gaps = 7/135 (5%)
Frame = +1
Query: 271 HHHTTVPGRR*LGPHLMHLCAHALCGILPLNP*RAISTAHAHKTSITSHPHTPQRHTRTP 450
HHH P GPH M L G++P P + S S P Q+ T P
Sbjct: 393 HHHWMQPHHS--GPHPMGLLHSHPMGLMPPPPPPSGPAPPPPPPSAPSLPPWQQQATGAP 450
Query: 451 HQLC-HLPITAASVTHASRLSRLPPLAGP*WPSPASAA------ASVAPCSPDAHASAGG 609
Q HLP +S T S + LPP + +S+A AS+ P P
Sbjct: 451 PQTGGHLPWHQSSPTTVSGTASLPPWQQGAGSTTSSSAPQLQTTASLVPPLPGVQP---- 506
Query: 610 PLPPAPVPAPPPPPP 654
PLPP+ P PPPPPP
Sbjct: 507 PLPPSAPPPPPPPPP 521
>ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca]
Length = 754
Score = 50.4 bits (119), Expect = 2e-05
Identities = 32/108 (29%), Positives = 44/108 (40%), Gaps = 12/108 (11%)
Frame = +1
Query: 355 PLNP*RAISTAHAHKTSITSHPHTPQRHTRTPHQLCHLPITAASVTHASRLSRLPPLAGP 534
P P R+ S + S + HP P + P + P T + AS S P P
Sbjct: 186 PPEPLRSASPSKNSTNSPSPHPPAPAASSSCPTSTANAPPTCPTPPAASTASPEPTSPPP 245
Query: 535 *WPSPASAAASVAPCSPD------------AHASAGGPLPPAPVPAPP 642
WP+P+S A S A +P + +SA P PP P+PP
Sbjct: 246 TWPAPSSKACSAASLTPSTPSSTKVSPSTVSSSSAAAPAPPQSAPSPP 293
Score = 35.8 bits (81), Expect = 0.61
Identities = 35/109 (32%), Positives = 44/109 (40%), Gaps = 8/109 (7%)
Frame = +1
Query: 355 PLNP*RAISTAHAHKTSITSHPHTPQRHTRTPHQLCHLPITAASVTHASRLSRLPPLAGP 534
P NP +T A T T+ P TP T +P S + PP P
Sbjct: 54 PPNPSPTAATPPAPAT--TTPPPTPTGPTCSPSPSAANSAPRESPPPPKSSAASPPPNSP 111
Query: 535 *WPSPASAAASV----APCSPDAHAS---AGGPLPPAPVPA-PPPPPPA 657
S A A A+ +P +PD S + P PP+P P PPP PPA
Sbjct: 112 PSTSSAPAPATTWPPHSPSTPDPATSSSPSAPPEPPSPSPKNPPPTPPA 160
Score = 34.3 bits (77), Expect = 1.8
Identities = 21/64 (32%), Positives = 30/64 (46%), Gaps = 1/64 (1%)
Frame = +1
Query: 469 PITAASVTHASRLSRLPPLAGP*WPSPASAAASVAPCSPDAHASAGGP-LPPAPVPAPPP 645
P + S + A + P P P PA++++ AP P + + P PPAP P P
Sbjct: 108 PNSPPSTSSAPAPATTWPPHSPSTPDPATSSSPSAPPEPPSPSPKNPPPTPPAPSPDSPT 167
Query: 646 PPPA 657
PP A
Sbjct: 168 PPDA 171
Score = 33.5 bits (75), Expect = 3.0
Identities = 19/69 (27%), Positives = 33/69 (47%)
Frame = +1
Query: 445 TPHQLCHLPITAASVTHASRLSRLPPLAGP*WPSPASAAASVAPCSPDAHASAGGPLPPA 624
TP ++ ++V+ +S + PP + P SP ++A+ +P P A+ S P
Sbjct: 261 TPSTPSSTKVSPSTVSSSSAAAPAPPQSAP---SPPKSSAAPSPSQPKANTSPTAPPAKQ 317
Query: 625 PVPAPPPPP 651
P +P PP
Sbjct: 318 PGHSPANPP 326
>emb|CAD70966.1| probable Cytokinesis protein sepA [Neurospora crassa]
Length = 1790
Score = 49.3 bits (116), Expect = 5e-05
Identities = 33/93 (35%), Positives = 42/93 (44%)
Frame = +1
Query: 376 ISTAHAHKTSITSHPHTPQRHTRTPHQLCHLPITAASVTHASRLSRLPPLAGP*WPSPAS 555
I+T +H + + P TP T TP + +T A+ A + PP P P P
Sbjct: 988 ITTGPSHPSMESQTPITPS-DTETPK----IQVTDATGPSAPAAASGPPPPPPP-PPPPP 1041
Query: 556 AAASVAPCSPDAHASAGGPLPPAPVPAPPPPPP 654
P +P AGGP PP P P PPPPPP
Sbjct: 1042 PPPGFLPGAPAPIPGAGGPPPPPPPPPPPPPPP 1074
Score = 37.7 bits (86), Expect = 0.16
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = +1
Query: 517 PPLAGP*WPSPASAAASVAPCSPDAHASAGGPLPPAPVPAPPPPP 651
PP P P P AP P AGGP PP P P PPP P
Sbjct: 1063 PPPPPPPPPPPPGGLPGAAPPMP----GAGGPPPPPPPPPPPPMP 1103
>ref|XP_327870.1| hypothetical protein [Neurospora crassa] gi|28917030|gb|EAA26755.1|
hypothetical protein [Neurospora crassa]
Length = 1817
Score = 49.3 bits (116), Expect = 5e-05
Identities = 33/93 (35%), Positives = 42/93 (44%)
Frame = +1
Query: 376 ISTAHAHKTSITSHPHTPQRHTRTPHQLCHLPITAASVTHASRLSRLPPLAGP*WPSPAS 555
I+T +H + + P TP T TP + +T A+ A + PP P P P
Sbjct: 988 ITTGPSHPSMESQTPITPS-DTETPK----IQVTDATGPSAPAAASGPPPPPPP-PPPPP 1041
Query: 556 AAASVAPCSPDAHASAGGPLPPAPVPAPPPPPP 654
P +P AGGP PP P P PPPPPP
Sbjct: 1042 PPPGFLPGAPAPIPGAGGPPPPPPPPPPPPPPP 1074
Score = 37.7 bits (86), Expect = 0.16
Identities = 20/45 (44%), Positives = 20/45 (44%)
Frame = +1
Query: 517 PPLAGP*WPSPASAAASVAPCSPDAHASAGGPLPPAPVPAPPPPP 651
PP P P P AP P AGGP PP P P PPP P
Sbjct: 1063 PPPPPPPPPPPPGGLPGAAPPMP----GAGGPPPPPPPPPPPPMP 1103
Score = 32.0 bits (71), Expect = 8.9
Identities = 20/54 (37%), Positives = 21/54 (38%), Gaps = 8/54 (14%)
Frame = +1
Query: 517 PPLAGP*WPSPASAAASVAPCSPDAHASAGGPLPPAPVPA--------PPPPPP 654
PP+ G P P P P AG P PP P P PPPPPP
Sbjct: 1082 PPMPGAGGPPPPPPP----PPPPPMPGMAGMPPPPPPPPPMPGMPGMPPPPPPP 1131