Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001362A_C01 KCC001362A_c01
(565 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
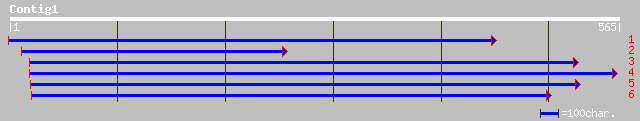
ref|ZP_00012769.1| COG0050: GTPases - translation elongation fac... 157 7e-38
ref|ZP_00012799.1| COG0050: GTPases - translation elongation fac... 157 7e-38
ref|ZP_00052061.1| COG0050: GTPases - translation elongation fac... 157 7e-38
ref|ZP_00013721.1| COG0050: GTPases - translation elongation fac... 155 4e-37
ref|ZP_00013734.1| COG0050: GTPases - translation elongation fac... 155 4e-37
>ref|ZP_00012769.1| COG0050: GTPases - translation elongation factors [Rhodopseudomonas
palustris]
Length = 396
Score = 157 bits (398), Expect = 7e-38
Identities = 74/87 (85%), Positives = 80/87 (91%)
Frame = +2
Query: 305 RTKPHLNVGTIGHVDHGKTTLTAAITKVLAETGGSTKVVAYDQIDKAPEEKARGITINAT 484
RTKPH N+GTIGHVDHGKT+LTAAITKVLAETGG+T AYDQIDKAPEEKARGITI+
Sbjct: 8 RTKPHCNIGTIGHVDHGKTSLTAAITKVLAETGGAT-FTAYDQIDKAPEEKARGITISTA 66
Query: 485 HVEYQTEKRHYAHVDCPGHADYVKNMI 565
HVEY+T+ RHYAHVDCPGHADYVKNMI
Sbjct: 67 HVEYETQNRHYAHVDCPGHADYVKNMI 93
>ref|ZP_00012799.1| COG0050: GTPases - translation elongation factors [Rhodopseudomonas
palustris]
Length = 419
Score = 157 bits (398), Expect = 7e-38
Identities = 74/87 (85%), Positives = 80/87 (91%)
Frame = +2
Query: 305 RTKPHLNVGTIGHVDHGKTTLTAAITKVLAETGGSTKVVAYDQIDKAPEEKARGITINAT 484
RTKPH N+GTIGHVDHGKT+LTAAITKVLAETGG+T AYDQIDKAPEEKARGITI+
Sbjct: 31 RTKPHCNIGTIGHVDHGKTSLTAAITKVLAETGGAT-FTAYDQIDKAPEEKARGITISTA 89
Query: 485 HVEYQTEKRHYAHVDCPGHADYVKNMI 565
HVEY+T+ RHYAHVDCPGHADYVKNMI
Sbjct: 90 HVEYETQNRHYAHVDCPGHADYVKNMI 116
>ref|ZP_00052061.1| COG0050: GTPases - translation elongation factors [Magnetospirillum
magnetotacticum]
Length = 425
Score = 157 bits (398), Expect = 7e-38
Identities = 74/88 (84%), Positives = 81/88 (91%)
Frame = +2
Query: 302 SRTKPHLNVGTIGHVDHGKTTLTAAITKVLAETGGSTKVVAYDQIDKAPEEKARGITINA 481
SRTKPH N+GTIGHVDHGKT+LTAAITKVLAE+GG+T AYDQIDKAPEEKARGITI+
Sbjct: 7 SRTKPHCNIGTIGHVDHGKTSLTAAITKVLAESGGAT-FTAYDQIDKAPEEKARGITIST 65
Query: 482 THVEYQTEKRHYAHVDCPGHADYVKNMI 565
HVEY+T+ RHYAHVDCPGHADYVKNMI
Sbjct: 66 AHVEYETQNRHYAHVDCPGHADYVKNMI 93
>ref|ZP_00013721.1| COG0050: GTPases - translation elongation factors [Rhodospirillum
rubrum]
Length = 396
Score = 155 bits (392), Expect = 4e-37
Identities = 74/88 (84%), Positives = 80/88 (90%)
Frame = +2
Query: 302 SRTKPHLNVGTIGHVDHGKTTLTAAITKVLAETGGSTKVVAYDQIDKAPEEKARGITINA 481
+RTKPH NVGTIGHVDHGKT+LTAAITKVLAE GG+T AYDQIDKAPEE+ARGITI+
Sbjct: 7 ARTKPHCNVGTIGHVDHGKTSLTAAITKVLAEAGGAT-FQAYDQIDKAPEERARGITIST 65
Query: 482 THVEYQTEKRHYAHVDCPGHADYVKNMI 565
HVEY+TE RHYAHVDCPGHADYVKNMI
Sbjct: 66 AHVEYETEARHYAHVDCPGHADYVKNMI 93
>ref|ZP_00013734.1| COG0050: GTPases - translation elongation factors [Rhodospirillum
rubrum]
Length = 447
Score = 155 bits (392), Expect = 4e-37
Identities = 74/88 (84%), Positives = 80/88 (90%)
Frame = +2
Query: 302 SRTKPHLNVGTIGHVDHGKTTLTAAITKVLAETGGSTKVVAYDQIDKAPEEKARGITINA 481
+RTKPH NVGTIGHVDHGKT+LTAAITKVLAE GG+T AYDQIDKAPEE+ARGITI+
Sbjct: 58 ARTKPHCNVGTIGHVDHGKTSLTAAITKVLAEAGGAT-FQAYDQIDKAPEERARGITIST 116
Query: 482 THVEYQTEKRHYAHVDCPGHADYVKNMI 565
HVEY+TE RHYAHVDCPGHADYVKNMI
Sbjct: 117 AHVEYETEARHYAHVDCPGHADYVKNMI 144