Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001337A_C01 KCC001337A_c01
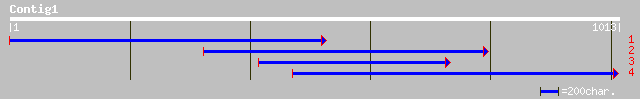
(1013 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S57795 probable deoxyribodipyrimidine photo-lyase (EC 4.1.9... 67 4e-10
dbj|BAA78564.1| amyR [Emericella nidulans] gi|6561865|gb|AAF1710... 36 1.2
ref|NP_726183.1| defective proventriculus CG5799-PC [Drosophila ... 35 2.7
pir||T00117 dve protein - fruit fly (Drosophila melanogaster) gi... 35 2.7
emb|CAA09729.1| Dve protein [Drosophila melanogaster] 35 2.7
>pir||S57795 probable deoxyribodipyrimidine photo-lyase (EC 4.1.99.3) -
Chlamydomonas reinhardtii
Length = 867
Score = 67.4 bits (163), Expect = 4e-10
Identities = 31/31 (100%), Positives = 31/31 (100%)
Frame = +3
Query: 48 EARSALQGGFFVAKRLRCSTNIHVYLVCMHA 140
EARSALQGGFFVAKRLRCSTNIHVYLVCMHA
Sbjct: 837 EARSALQGGFFVAKRLRCSTNIHVYLVCMHA 867
>dbj|BAA78564.1| amyR [Emericella nidulans] gi|6561865|gb|AAF17101.1|AF208225_1
amylase cluster transcriptional regulator AmyR
[Emericella nidulans]
Length = 662
Score = 35.8 bits (81), Expect = 1.2
Identities = 31/104 (29%), Positives = 41/104 (38%), Gaps = 16/104 (15%)
Frame = -2
Query: 916 HQATPTFLRSMSENCSSKRTQDMSGAWPAA-------SPSMCAQCSAAW---------RE 785
HQ P L S S + + T D+SG WPAA S S AW +
Sbjct: 562 HQINPPLLNSASSSSIASETLDISGPWPAADDNTMDSSTGSRTTNSCAWEITSGDTNDQN 621
Query: 784 NASTRPPAPYLFAAVLLCAPLFLRQRYHSQPPFIAVLTFSAVKL 653
S +PP+ ++V L R HS A L+F A +L
Sbjct: 622 QTSDQPPSLSGSSSVPLSV-----SRMHSSTSMAAELSFQAGRL 660
>ref|NP_726183.1| defective proventriculus CG5799-PC [Drosophila melanogaster]
gi|21626548|gb|AAM68215.1| CG5799-PC [Drosophila
melanogaster]
Length = 1039
Score = 34.7 bits (78), Expect = 2.7
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Frame = +3
Query: 720 NKGAHNSTAAKRYGAG--GRVLAFSLHAALHWAHMEGEAAGHAP 845
N+ AH++ A + G G G +L +HA++H H G GH P
Sbjct: 522 NEAAHHAGAGQAPGPGHHGSMLVHPMHASMHHHHHHGGGGGHGP 565
>pir||T00117 dve protein - fruit fly (Drosophila melanogaster)
gi|3523073|dbj|BAA32660.1| Dve [Drosophila melanogaster]
Length = 1019
Score = 34.7 bits (78), Expect = 2.7
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Frame = +3
Query: 720 NKGAHNSTAAKRYGAG--GRVLAFSLHAALHWAHMEGEAAGHAP 845
N+ AH++ A + G G G +L +HA++H H G GH P
Sbjct: 502 NEAAHHAGAGQAPGPGHHGSMLVHPMHASMHHHHHHGGGGGHGP 545
>emb|CAA09729.1| Dve protein [Drosophila melanogaster]
Length = 1021
Score = 34.7 bits (78), Expect = 2.7
Identities = 16/44 (36%), Positives = 24/44 (54%), Gaps = 2/44 (4%)
Frame = +3
Query: 720 NKGAHNSTAAKRYGAG--GRVLAFSLHAALHWAHMEGEAAGHAP 845
N+ AH++ A + G G G +L +HA++H H G GH P
Sbjct: 504 NEAAHHAGAGQAPGPGHHGSMLVHPMHASMHHHHHHGGGGGHGP 547