Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001333A_C01 KCC001333A_c01
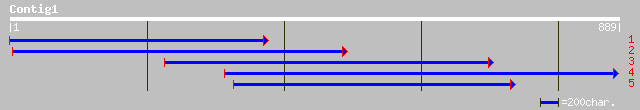
(889 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|ZP_00025308.1| COG1012: NAD-dependent aldehyde dehydrogenase... 37 0.001
ref|XP_286444.1| hypothetical protein XP_286444 [Mus musculus] 44 0.004
ref|NP_787118.1| hypothetical protein MGC35308 [Homo sapiens] gi... 42 0.014
ref|NP_079362.1| hypothetical protein FLJ22596 [Homo sapiens] gi... 42 0.014
emb|CAB98161.1| hypothetical protein L1375.07 [Leishmania major] 42 0.014
>ref|ZP_00025308.1| COG1012: NAD-dependent aldehyde dehydrogenases [Ralstonia
metallidurans]
Length = 626
Score = 37.0 bits (84), Expect(2) = 0.001
Identities = 35/116 (30%), Positives = 42/116 (36%), Gaps = 16/116 (13%)
Frame = -2
Query: 885 HAHAHAHAHTHPGHSSKAAPKSRGKASFRT-----QPPRPATFPQSRWRPRRRPAH---- 733
H H H GH AA + R S QP RPA S R A
Sbjct: 68 HGGGHLHGARPCGHGRAAAGRVRAAGSHAARPVPGQPLRPALLASSATGDARMKAQEIAA 127
Query: 732 ------CPPP-QPASTNHPSSVNIRQHLCT*GQSYPLFCQCAAAVANARHTRADTA 586
PPP PA P + +R + G+ + C AA+ANAR RA A
Sbjct: 128 CWQAMDLPPPWTPAEGAAPGVLAVRSPID--GEVFGQLAMCDAALANARIARAHAA 181
Score = 27.7 bits (60), Expect(2) = 0.001
Identities = 16/39 (41%), Positives = 21/39 (53%)
Frame = -1
Query: 502 AHALPLLSALIPAKSRGLVCPKRGVLFRQNQTAHARLMT 386
AHA AL PA +RG V + G R N+ A RL++
Sbjct: 178 AHAAQTTWALTPAPARGEVVRRFGEALRANKDALGRLVS 216
>ref|XP_286444.1| hypothetical protein XP_286444 [Mus musculus]
Length = 95
Score = 43.9 bits (102), Expect = 0.004
Identities = 24/54 (44%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Frame = -3
Query: 887 HTHTHTHTHTHTPVIAARPLPKAEGRRHFALSLHDRP-HSPRAAGAHAGALPIA 729
HTHTHTHTHTH+PV A + + HD P HS AH LP A
Sbjct: 23 HTHTHTHTHTHSPVSAIIAFQSPKSSPLCKMRSHDLPAHSSPLNPAHFYMLPAA 76
>ref|NP_787118.1| hypothetical protein MGC35308 [Homo sapiens]
gi|21961551|gb|AAH34775.1| Hypothetical protein MGC35308
[Homo sapiens]
Length = 295
Score = 42.0 bits (97), Expect = 0.014
Identities = 22/52 (42%), Positives = 26/52 (49%), Gaps = 1/52 (1%)
Frame = -2
Query: 852 PGHSSKAAPKSRGKASFRTQPPRP-ATFPQSRWRPRRRPAHCPPPQPASTNH 700
P + K P RG RTQPP P PQ+ R PA C PP+PA + H
Sbjct: 49 PSATLKRPPARRGPGLDRTQPPAPPGVSPQALPSRARAPATCAPPRPAGSGH 100
>ref|NP_079362.1| hypothetical protein FLJ22596 [Homo sapiens]
gi|10439043|dbj|BAB15412.1| unnamed protein product
[Homo sapiens]
Length = 160
Score = 42.0 bits (97), Expect = 0.014
Identities = 17/24 (70%), Positives = 18/24 (74%)
Frame = +1
Query: 817 SAFGSGLAAMTGVCVCVCVCVCVC 888
S G GL + GVCVCVCVCVCVC
Sbjct: 23 SPLGPGLDWVCGVCVCVCVCVCVC 46
>emb|CAB98161.1| hypothetical protein L1375.07 [Leishmania major]
Length = 1092
Score = 42.0 bits (97), Expect = 0.014
Identities = 43/171 (25%), Positives = 62/171 (36%), Gaps = 8/171 (4%)
Frame = -3
Query: 803 FALSLHDRPHSPRAAGAHAGALPIAHLPNQQAPITHHP--------*TSANIYALEVKAI 648
FA S RP AG +G+ P H+P+ +P H P +S + V +
Sbjct: 85 FASSPEVRPSFSTRAGGPSGSNPAPHMPSVSSPHGHRPHDGGVSSASSSGQPVSSSVDRL 144
Query: 647 LCFVSARQQWLMRGTHARTQRQNAQSNAGKQAAPTPAAPPALGSDGLSSTRPPFALRLDS 468
Q R +HA R+ + + PTPA A P +RL
Sbjct: 145 AALYQQFQMSQRRDSHA--PRRGTAGRSREAVIPTPARQEA----------PAAPVRLSR 192
Query: 467 GKEQRISVSQAWSAVPSKPNRPCTAHDSAAPRFHPRISRDQAGQKGVGTHL 315
+ + + + KPNR S+AP PR A KG G H+
Sbjct: 193 QTSRGAASGDSGATDKEKPNRIAPGGGSSAPARVPR-----AASKGGGAHV 238