Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001328A_C01 KCC001328A_c01
(724 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
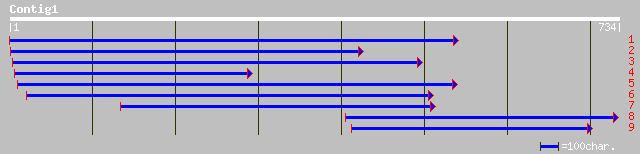
emb|CAD38626.1| hypothetical protein [Homo sapiens] 36 0.53
ref|XP_316196.1| ENSANGP00000005931 [Anopheles gambiae] gi|30175... 35 0.90
ref|XP_317879.1| ENSANGP00000004917 [Anopheles gambiae] gi|21300... 35 0.90
ref|ZP_00071800.1| COG0465: ATP-dependent Zn proteases [Trichode... 35 1.2
ref|NP_859505.1| hypothetical protein Chr3_0720 [Leishmania majo... 34 2.0
>emb|CAD38626.1| hypothetical protein [Homo sapiens]
Length = 217
Score = 36.2 bits (82), Expect = 0.53
Identities = 23/52 (44%), Positives = 23/52 (44%), Gaps = 8/52 (15%)
Frame = +1
Query: 196 GQPPSMPPALGRRRKPR--WLLRPCPGPRGLSW------ATGCVNLTCSSAS 327
G PPS PA R R W L CPG R SW G V TCSS S
Sbjct: 35 GAPPSPAPAPARPRPALCIWPLGSCPGRRPASWGRAMSHGAGLVRTTCSSGS 86
>ref|XP_316196.1| ENSANGP00000005931 [Anopheles gambiae] gi|30175949|gb|EAA10839.2|
ENSANGP00000005931 [Anopheles gambiae str. PEST]
Length = 2680
Score = 35.4 bits (80), Expect = 0.90
Identities = 23/70 (32%), Positives = 36/70 (50%), Gaps = 1/70 (1%)
Frame = -2
Query: 273 GARAWSKQPSRLAASSERG-RHARGLPPAGRQRPPR*AESGD*MSACR*QPSRCGAEKAD 97
GA A +K+P+ AAS++RG +HA +G R +G S+ GA+K +
Sbjct: 2399 GANAATKEPTGAAASAKRGQKHATPAAKSGNARSSGGRGAGQ---------SKRGAKKNN 2449
Query: 96 RKSTFCFCET 67
+ T C C+T
Sbjct: 2450 KAQTHCICQT 2459
>ref|XP_317879.1| ENSANGP00000004917 [Anopheles gambiae] gi|21300834|gb|EAA12979.1|
ENSANGP00000004917 [Anopheles gambiae str. PEST]
Length = 257
Score = 35.4 bits (80), Expect = 0.90
Identities = 48/170 (28%), Positives = 56/170 (32%), Gaps = 22/170 (12%)
Frame = -2
Query: 612 RRSHVPGVCSP----------HGPPRKKTCALGGQPRSARHQS*ARGPARGILLD*PRST 463
RR P C P H PPR + CA AR A GP R ST
Sbjct: 93 RRPRSPPFCFPPPAAAPARRAHRPPRCR-CAPAAPSPPAR----AFGPWRS-------ST 140
Query: 462 RGQGAAP*PYERPGEIHTCGWRGRHHDPHRSC*SH*SQT------ATVCRATC*RTGQVD 301
R +P + T GWR P R C + S T T CR T T
Sbjct: 141 RSSARSP-------QSPTAGWRSFAPTPRRPCTAADSGTRRTAASGTRCRGTPSGTRCSA 193
Query: 300 AASCPAQPPGARAWSKQPSRLAASSERGRHARGLPPAGRQR------PPR 169
A C A P AR+ P ++S + P R PPR
Sbjct: 194 PAPCAASPASARSPPVSPRTARSASRPSSRSASAPSRASPRTCSCRPPPR 243
>ref|ZP_00071800.1| COG0465: ATP-dependent Zn proteases [Trichodesmium erythraeum
IMS101]
Length = 667
Score = 35.0 bits (79), Expect = 1.2
Identities = 19/63 (30%), Positives = 33/63 (52%), Gaps = 2/63 (3%)
Frame = +3
Query: 150 NRQTRLNVVAFGGQQGAAPEHAARARTTPQASMAASTM--PGPQGAELGNWLRQLDLFFS 323
+RQ +++ A+ G+ G HA + TP+ S+ A PG GA+L N L + + +
Sbjct: 365 DRQVTVDLPAYKGRLGILEVHARNKKLTPEISLEAIARKTPGFSGADLANMLNEAAILTA 424
Query: 324 KSR 332
+ R
Sbjct: 425 RRR 427
>ref|NP_859505.1| hypothetical protein Chr3_0720 [Leishmania major]
gi|21629369|gb|AAM69046.1|AC125735_76 hypothetical
protein Chr3_0720 [Leishmania major]
Length = 1078
Score = 34.3 bits (77), Expect = 2.0
Identities = 23/79 (29%), Positives = 33/79 (41%)
Frame = -3
Query: 326 LAEEQVKLTQPVAQLSPLGPGHGRSSHRGLRRRPSAGGMLGGCPLLAAKGHHVEPSLAIR 147
L E Q L +SP GPG G S + L R S GG + G + A G H + ++
Sbjct: 741 LGEAQYTLPPATMLMSPRGPGGGASRYHPLHRN-SEGGNVAGSAVTDAPGRHRQQQYWLQ 799
Query: 146 *ALVGSSRHAAALKKQTGS 90
H A ++ G+
Sbjct: 800 GGFALHGDHPAVAQRGGGA 818