Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001327A_C01 KCC001327A_c01
(807 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
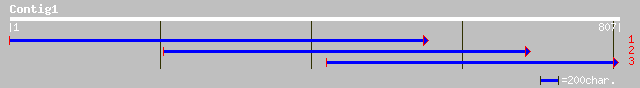
ref|XP_316558.1| ENSANGP00000010090 [Anopheles gambiae] gi|21299... 57 4e-07
ref|NP_767161.1| blr0521 [Bradyrhizobium japonicum] gi|27348769|... 55 2e-06
ref|ZP_00054604.1| COG0659: Sulfate permease and related transpo... 55 2e-06
emb|CAB62280.1| hydroxyproline-rich glycoprotein DZ-HRGP [Volvox... 54 2e-06
ref|XP_233556.2| similar to Ser/Arg-related nuclear matrix prote... 54 2e-06
>ref|XP_316558.1| ENSANGP00000010090 [Anopheles gambiae] gi|21299146|gb|EAA11291.1|
ENSANGP00000010090 [Anopheles gambiae str. PEST]
Length = 536
Score = 57.0 bits (136), Expect = 4e-07
Identities = 37/114 (32%), Positives = 52/114 (45%), Gaps = 5/114 (4%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTP---- 176
P + TP+P C P PPP + P + PP++ P ++ A C+ P
Sbjct: 128 PYVATTTPAPP-------CARPTNPPPRYAPQPPQNYAPPKYAP--YQPPATCAPPPVQY 178
Query: 177 -RRPLTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPSTRCS 335
P T APP+A P P P P S+TPP A P P + TA P ++ C+
Sbjct: 179 TAPPTTCAPPTAPPRPYYQPSPPPSYTPPKYLPAPPPPPYSYTAPPRGEASTCA 232
Score = 39.3 bits (90), Expect = 0.076
Identities = 33/108 (30%), Positives = 43/108 (39%), Gaps = 5/108 (4%)
Frame = +3
Query: 3 TRPLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*-----SCLPPRWTPTPWRRRAPC 167
T P P + PSP + + P PPP + P +C PP+ + P APC
Sbjct: 189 TAPPRPYYQPSPP-PSYTPPKYLPAPPPPPYSYTAPPRGEASTCAPPKPSYLP----APC 243
Query: 168 STPRRPLTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACP 311
+ P P PP C S + PPS + P P P A T P
Sbjct: 244 NGPPPP---PPPPPCGSSVTMQPPSYQYQPTPPPAPKPYTYPATTPQP 288
>ref|NP_767161.1| blr0521 [Bradyrhizobium japonicum] gi|27348769|dbj|BAC45786.1|
blr0521 [Bradyrhizobium japonicum USDA 110]
Length = 745
Score = 54.7 bits (130), Expect = 2e-06
Identities = 37/92 (40%), Positives = 44/92 (47%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAA 197
P+ TP+P T + P APPP R P + P P P AP +TP TA
Sbjct: 234 PTATPAPGSTPGAPPAGRPGAPPPGVRPGSPPAAGSP---PAPGATPAPTTTPAPGGTAT 290
Query: 198 PPSACPSPASSPPPSRSWTPPTTPRAAPAGPL 293
PPS P PAS+P P + TP T AP G L
Sbjct: 291 PPSGRPGPASTPAPGAA-TPAPTATPAPGGAL 321
Score = 50.8 bits (120), Expect = 3e-05
Identities = 38/109 (34%), Positives = 45/109 (40%), Gaps = 2/109 (1%)
Frame = +3
Query: 6 RPLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRP 185
RP P P PA +P P P P + P + PTP P + P
Sbjct: 105 RPAPPPPPPPPA---------APKQPSP------PPAAAPQQHAPTPPPPAPPAARPAPT 149
Query: 186 LTAAPPSACPSPASSPPPSRSWTP-PTTPRAAPAGPLAR-TACPSCPST 326
A PP+A P A PPP + P PT P PAGP AR T P+ T
Sbjct: 150 PPAPPPAAAPQHAPPPPPPPAARPTPTPPPPPPAGPAARPTPAPTATPT 198
Score = 48.9 bits (115), Expect = 1e-04
Identities = 44/126 (34%), Positives = 51/126 (39%), Gaps = 21/126 (16%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLP-----------PRWTPTPWRR 155
P P+ P+PA T +P+APPPA T P S P P TP P
Sbjct: 182 PAGPAARPTPAPTATP----TPVAPPPAAPTARPGSPAPAATPAPTPTPAPTATPAPTAT 237
Query: 156 RAPCSTPRRPLT----AAPPSACPS--PASSPPPSRSWTPPTTPRAAPAG----PLARTA 305
AP STP P A PP P PA+ PP+ TP T AP G P R
Sbjct: 238 PAPGSTPGAPPAGRPGAPPPGVRPGSPPAAGSPPAPGATPAPTTTPAPGGTATPPSGRPG 297
Query: 306 CPSCPS 323
S P+
Sbjct: 298 PASTPA 303
Score = 43.9 bits (102), Expect = 0.003
Identities = 24/66 (36%), Positives = 31/66 (46%)
Frame = +3
Query: 123 PPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLART 302
PP+ P AP + P RP P +A P P ++PPP+ + P P P P AR
Sbjct: 51 PPKGPPGA----APPAAPARPAAPPPAAAPPHPPAAPPPAAAPPRPAAPPPPPPPPAARP 106
Query: 303 ACPSCP 320
A P P
Sbjct: 107 APPPPP 112
Score = 43.1 bits (100), Expect = 0.005
Identities = 29/73 (39%), Positives = 31/73 (41%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P APPPA P PP P P R AP P P AAP P PA++P
Sbjct: 80 PAAPPPAAAP--PRPAAPPPPPPPPAARPAPPPPPPPP--AAPKQPSPPPAAAPQQHAPT 135
Query: 252 TPPTTPRAAPAGP 290
PP P AA P
Sbjct: 136 PPPPAPPAARPAP 148
Score = 42.4 bits (98), Expect = 0.009
Identities = 35/103 (33%), Positives = 43/103 (40%), Gaps = 1/103 (0%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPL 188
P P+ P+P +P APPPA P PP P P R P P P
Sbjct: 139 PAPPAARPAP----------TPPAPPPAAA---PQHAPPP--PPPPAARPTPTPPPPPP- 182
Query: 189 TAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPL-ARTACPS 314
A P+A P+PA + P+ PP P A P P A T P+
Sbjct: 183 --AGPAARPTPAPTATPTPVAPPPAAPTARPGSPAPAATPAPT 223
Score = 41.2 bits (95), Expect = 0.020
Identities = 28/83 (33%), Positives = 32/83 (37%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P APPPA PP P AP P PP+A P+P PPP +
Sbjct: 67 PAAPPPA--------AAPPHPPAAPPPAAAPPRPAAPPPPPPPPAARPAPPPPPPPPAAP 118
Query: 252 TPPTTPRAAPAGPLARTACPSCP 320
P+ P AA A T P P
Sbjct: 119 KQPSPPPAAAPQQHAPTPPPPAP 141
Score = 40.0 bits (92), Expect = 0.044
Identities = 23/55 (41%), Positives = 26/55 (46%), Gaps = 1/55 (1%)
Frame = +3
Query: 162 PCSTPRRPLTAAPPSACPS-PASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPS 323
P + P P A PP+A P PA+ PPP PP R AP P A P PS
Sbjct: 72 PAAAPPHPPAAPPPAAAPPRPAAPPPP----PPPPAARPAPPPPPPPPAAPKQPS 122
Score = 40.0 bits (92), Expect = 0.044
Identities = 37/113 (32%), Positives = 44/113 (38%), Gaps = 12/113 (10%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPP----AWRT**P*SCLPPRWTPTPWRRRAPCST---- 173
P+ TP+P L PPP A T P PP P AP
Sbjct: 311 PTATPAPG---------GALTPPPGRPGAGPTPGPQGGTPPAGAPAAGTPAAPPQAGGLP 361
Query: 174 --PRRPLTAAPPSACP-SPASSPPPSRS-WTPPTTPRAAPAGPLARTACPSCP 320
P P AA PS P S A++PPP+R+ + PPT A A P P P
Sbjct: 362 ARPAAPAGAAAPSTVPGSAAATPPPNRAQFAPPTVTPAFQAAPTVVAPLPPPP 414
Score = 38.9 bits (89), Expect = 0.099
Identities = 23/58 (39%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Frame = +3
Query: 162 PCSTPRRPLTAAPPSACPSPASSPPPSRSWTPPTT--PRAAPAGPLARTACPSCPSTR 329
P P+ P AAPP+A PA+ PP + PP P AAP P A P P+ R
Sbjct: 48 PKQPPKGPPGAAPPAAPARPAAPPPAAAPPHPPAAPPPAAAPPRPAAPPPPPPPPAAR 105
Score = 38.9 bits (89), Expect = 0.099
Identities = 32/91 (35%), Positives = 39/91 (42%), Gaps = 8/91 (8%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPP--------RWTPTPWRRRAPCSTPRRPLTAAPPSACPSPAS 227
P PPPA R P PP R TP P P + P TA P S P+PA+
Sbjct: 164 PPPPPPAAR---PTPTPPPPPPAGPAARPTPAPTATPTPVAPPPAAPTARPGS--PAPAA 218
Query: 228 SPPPSRSWTPPTTPRAAPAGPLARTACPSCP 320
+P P+ + P TP AP A + P P
Sbjct: 219 TPAPTPTPAPTATP--APTATPAPGSTPGAP 247
Score = 33.1 bits (74), Expect = 5.4
Identities = 31/107 (28%), Positives = 37/107 (33%), Gaps = 2/107 (1%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRR-APCSTPRRP 185
P P TP+P T A PP+ R P TP P AP +TP
Sbjct: 270 PPAPGATPAPTTTPAPG----GTATPPSGRP-------GPASTPAPGAATPAPTATPAPG 318
Query: 186 LTAAPPSACPSPASSPPPSRSWTPPTTPRA-APAGPLARTACPSCPS 323
PP P +P P P P A PA P P+ P+
Sbjct: 319 GALTPPPGRPGAGPTPGPQGGTPPAGAPAAGTPAAPPQAGGLPARPA 365
>ref|ZP_00054604.1| COG0659: Sulfate permease and related transporters (MFS
superfamily) [Magnetospirillum magnetotacticum]
Length = 1205
Score = 54.7 bits (130), Expect = 2e-06
Identities = 55/175 (31%), Positives = 69/175 (39%), Gaps = 28/175 (16%)
Frame = -3
Query: 445 ANAGVARR----HAQHAGQEEAQ-------HGGVRRRLQDHAAHAQHDEHRVDGQLGQAV 299
A GV RR H G E +Q HGG R+ L A + EHR +GQA
Sbjct: 129 AGRGVDRRCGDRDRHHGGLEGSQPVGDGDAHGGARQGLLQRAGRNGNLEHRPAPAIGQAD 188
Query: 298 LANGPAGAARGVVGGVHDLEGGGDDAGEGHADGGAAVRGLLGVEHGA------------- 158
G G+ GV +EGG G+ AA GL GV+ A
Sbjct: 189 AGLAEGGHVLGLHEGVGGIEGGLGRYGQPQHRAVAAAHGLCGVQGQAVAAAVAGDQGGKA 248
Query: 157 --LRRQGVGVHRGGRQDQG--HHVRHAGGGASGLQQQAEQHVQAGDGVNDGVSGL 5
+ +G G+ R GRQ Q H +RHA G E H Q G + G+ GL
Sbjct: 249 PDIMGEGSGLGRAGRQVQAQLHLLRHANLGTG-----QEVHHQMGGHRSCGIGGL 298
>emb|CAB62280.1| hydroxyproline-rich glycoprotein DZ-HRGP [Volvox carteri f.
nagariensis]
Length = 409
Score = 54.3 bits (129), Expect = 2e-06
Identities = 39/107 (36%), Positives = 46/107 (42%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPL 188
P +PS PSP P +PPP+ PP P+P P S P P
Sbjct: 147 PPSPSPPPSP-----------PPSPPPS---------PPPSPPPSP-----PPSPPPSPP 181
Query: 189 TAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPSTR 329
+ PP PSP SPPPS +PP P +P P R PS PS R
Sbjct: 182 PSPPPRPPPSPPPSPPPSPPPSPPPRPPPSPNPPSPRPPSPSPPSPR 228
Score = 46.6 bits (109), Expect = 5e-04
Identities = 31/83 (37%), Positives = 35/83 (41%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P PPP P PP P P P S P P + PPS PSP SPPPS
Sbjct: 123 PSPPPPPPPPPPPPPNPPPPPPPPPPSPSPPPSPPPSPPPSPPPSPPPSPPPSPPPSPPP 182
Query: 252 TPPTTPRAAPAGPLARTACPSCP 320
+PP P +P + PS P
Sbjct: 183 SPPPRPPPSPPPSPPPSPPPSPP 205
Score = 46.2 bits (108), Expect = 6e-04
Identities = 27/73 (36%), Positives = 30/73 (40%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P PPP+ P PP +P P R P P RP +P P P SPPP
Sbjct: 319 PRPPPPS-----PNPPRPPPPSPPPPRPPPPSPNPPRPPPPSPSPPKPPPPPSPPPRPPR 373
Query: 252 TPPTTPRAAPAGP 290
PP PR P P
Sbjct: 374 RPPRPPRRPPTAP 386
Score = 44.3 bits (103), Expect = 0.002
Identities = 34/104 (32%), Positives = 40/104 (37%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPL 188
P +PS P P P PPP P PP +P P P P P
Sbjct: 82 PPSPSPPPPPPPPVPPPPPPPPPPPPPPSPPPPPPPPPPPPPSPPPPPPPPPPPPPNPPP 141
Query: 189 TAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCP 320
PP PSP SPPPS +PP +P +P + PS P
Sbjct: 142 PPPPPPPSPSPPPSPPPSPPPSPPPSPPPSPPPSPPPSPPPSPP 185
Score = 44.3 bits (103), Expect = 0.002
Identities = 33/85 (38%), Positives = 37/85 (42%), Gaps = 1/85 (1%)
Frame = +3
Query: 69 SPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRS 248
SP PPP P PP P P P P P + PPS PSP SPPPS
Sbjct: 110 SPPPPPPPPPPPPPSPPPPPPPPPPPPPNPPPPPPPPPPSPSPPPSPPPSPPPSPPPSPP 169
Query: 249 WTPPTTPR-AAPAGPLARTACPSCP 320
+PP +P + P P R PS P
Sbjct: 170 PSPPPSPPPSPPPSPPPRPP-PSPP 193
Score = 44.3 bits (103), Expect = 0.002
Identities = 34/89 (38%), Positives = 40/89 (44%), Gaps = 4/89 (4%)
Frame = +3
Query: 69 SPLAP-PPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSP---P 236
SP +P PP R P PP +P P R P P RP +PP P P S P P
Sbjct: 223 SPPSPRPPPRRPPFP----PP--SPPPPRPPPPAPPPPRPPPPSPPPPLPPPPSPPPPLP 276
Query: 237 PSRSWTPPTTPRAAPAGPLARTACPSCPS 323
P S PP+ P +P P R PS P+
Sbjct: 277 PPPSPPPPSPPPPSPLPPSPRPPKPSPPN 305
Score = 42.4 bits (98), Expect = 0.009
Identities = 33/104 (31%), Positives = 40/104 (37%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPL 188
PL P +P P PL PPP+ P PP +P P R P +P
Sbjct: 264 PLPPPPSPPP-----------PLPPPPS-----PPPPSPPPPSPLPPSPRPPKPSPPNNP 307
Query: 189 TAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCP 320
PP P P PPPS + PP P +P P P+ P
Sbjct: 308 FPRPPPPNPPPPRPPPPSPN--PPRPPPPSPPPPRPPPPSPNPP 349
Score = 41.6 bits (96), Expect = 0.015
Identities = 35/104 (33%), Positives = 41/104 (38%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAA 197
PS PSP P +PPP+ P PP P+P P PR P +
Sbjct: 170 PSPPPSP-----------PPSPPPS-----PPPRPPPSPPPSPPPSPPPSPPPRPPPSPN 213
Query: 198 PPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPSTR 329
PPS P P S PPS P P P+ P R P+ P R
Sbjct: 214 PPS--PRPPSPSPPSPRPPPRRPPFPPPSPPPPRPPPPAPPPPR 255
Score = 41.6 bits (96), Expect = 0.015
Identities = 37/107 (34%), Positives = 42/107 (38%), Gaps = 5/107 (4%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRR---PL 188
PS PSP P +PPP+ P S PP P +P PRR P
Sbjct: 190 PSPPPSP-----------PPSPPPSPPPRPPPSPNPPSPRPPSPSPPSPRPPPRRPPFPP 238
Query: 189 TAAPPSACPSPASSP--PPSRSWTPPTTPRAAPAGPLARTACPSCPS 323
+ PP P PA P PP S PP P +P PL P PS
Sbjct: 239 PSPPPPRPPPPAPPPPRPPPPSPPPPLPPPPSPPPPLPPPPSPPPPS 285
Score = 40.8 bits (94), Expect = 0.026
Identities = 36/106 (33%), Positives = 39/106 (35%), Gaps = 4/106 (3%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPR----RP 185
PS PSP P PPP+ P P +P P RR P P RP
Sbjct: 198 PSPPPSP-----------PPRPPPSPNPPSPRPPSPSPPSPRPPPRRPPFPPPSPPPPRP 246
Query: 186 LTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPS 323
APP P P S PPP PP+ P P P P PS
Sbjct: 247 PPPAPPPPRPPPPSPPPPLPP--PPSPPPPLPPPPSPPPPSPPPPS 290
Score = 40.0 bits (92), Expect = 0.044
Identities = 33/103 (32%), Positives = 36/103 (34%), Gaps = 1/103 (0%)
Frame = +3
Query: 18 PSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAA 197
P+ P P + A +P APPP PP TP P P TP P
Sbjct: 20 PARKPPPRRSPVVALVETPAAPPPG---------SPPPGTPPPG---VPPPTPSGPEHPP 67
Query: 198 PPSACPSPASSPP-PSRSWTPPTTPRAAPAGPLARTACPSCPS 323
PP P P PP P PP P P P P PS
Sbjct: 68 PPPPPPPPPPQPPLPPSPSPPPPPPPPVPPPPPPPPPPPPPPS 110
Score = 39.7 bits (91), Expect = 0.058
Identities = 30/80 (37%), Positives = 33/80 (40%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P PPP P PP +P P R P P RP PPS P+P PPPS S
Sbjct: 309 PRPPPPN-----PPPPRPPPPSPNPPRPPPPSPPPPRP---PPPS--PNPPRPPPPSPSP 358
Query: 252 TPPTTPRAAPAGPLARTACP 311
P P + P P R P
Sbjct: 359 PKPPPPPSPPPRPPRRPPRP 378
Score = 38.9 bits (89), Expect = 0.099
Identities = 34/106 (32%), Positives = 36/106 (33%)
Frame = +3
Query: 6 RPLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRP 185
RP P +P P P PPPA P PP P P P P P
Sbjct: 233 RPPFPPPSPPP-----------PRPPPPAPPPPRPPPPSPPPPLPPPPSPPPPLPPPPSP 281
Query: 186 LTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPS 323
+PP P P S PP S PP P P P P PS
Sbjct: 282 PPPSPPPPSPLPPSPRPPKPS--PPNNPFPRPPPPNPPPPRPPPPS 325
Score = 38.5 bits (88), Expect = 0.13
Identities = 29/84 (34%), Positives = 31/84 (36%)
Frame = +3
Query: 72 PLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSW 251
P PPP + P S PP P P P P P + PP P P PPPS
Sbjct: 70 PPPPPPPPQPPLPPSPSPPPPPPPPVPPPPPPPPPPPPPPSPPPP--PPPPPPPPPSPPP 127
Query: 252 TPPTTPRAAPAGPLARTACPSCPS 323
PP P P P P PS
Sbjct: 128 PPPPPPPPPPNPPPPPPPPPPSPS 151
Score = 36.6 bits (83), Expect = 0.49
Identities = 34/115 (29%), Positives = 43/115 (36%), Gaps = 8/115 (6%)
Frame = +3
Query: 9 PLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTP--TPWRRRAPCSTPR- 179
PL P +P P P PPP+ P S PP+ +P P+ R P + P
Sbjct: 274 PLPPPPSPPP-----------PSPPPPSPL---PPSPRPPKPSPPNNPFPRPPPPNPPPP 319
Query: 180 -----RPLTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLARTACPSCPSTR 329
P PP P P PPPS + P P +P P + P P R
Sbjct: 320 RPPPPSPNPPRPPPPSPPPPRPPPPSPNPPRPPPPSPSPPKPPPPPSPPPRPPRR 374
Score = 33.1 bits (74), Expect = 5.4
Identities = 20/66 (30%), Positives = 23/66 (34%)
Frame = +3
Query: 123 PPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSWTPPTTPRAAPAGPLART 302
PP P P + P P PP P P PPP +PP P P P +
Sbjct: 67 PPPPPPPPPPPQPPLPPSPSPPPPPPPPVPPPPPPPPPPPPPPSPPPPPPPPPPPPPSPP 126
Query: 303 ACPSCP 320
P P
Sbjct: 127 PPPPPP 132
>ref|XP_233556.2| similar to Ser/Arg-related nuclear matrix protein;
plenty-of-prolines-101; serine/arginine repetitive
matrix protein 1 [Rattus norvegicus]
Length = 958
Score = 54.3 bits (129), Expect = 2e-06
Identities = 37/89 (41%), Positives = 44/89 (48%), Gaps = 5/89 (5%)
Frame = +3
Query: 78 APPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPP-SACPSPAS-SPPPSRSW 251
+PPPA R P PP P P RRR+P PRR + PP PSP SPP R +
Sbjct: 599 SPPPARRRRSPSPAPPPPPPPPPRRRRSPTPPPRRRTPSPPPRRRSPSPRRYSPPIQRRY 658
Query: 252 TPPTTPR---AAPAGPLARTACPSCPSTR 329
+P P+ A+P P R A PS P R
Sbjct: 659 SPSPPPKRRTASPPPPPKRRASPSPPPKR 687
Score = 40.4 bits (93), Expect = 0.034
Identities = 29/81 (35%), Positives = 38/81 (46%), Gaps = 13/81 (16%)
Frame = +3
Query: 81 PPPAWRT**P*----SCLPPRWTPTPWRRRAPCSTPRRPLTAAPP----SACPSP----- 221
PPP RT P S P R++P RR +P P+R + PP A PSP
Sbjct: 629 PPPRRRTPSPPPRRRSPSPRRYSPPIQRRYSPSPPPKRRTASPPPPPKRRASPSPPPKRR 688
Query: 222 ASSPPPSRSWTPPTTPRAAPA 284
S PP + +PP T R +P+
Sbjct: 689 VSHSPPPKQRSPPVTKRRSPS 709
Score = 38.9 bits (89), Expect = 0.099
Identities = 36/109 (33%), Positives = 50/109 (45%), Gaps = 7/109 (6%)
Frame = +3
Query: 135 TPTPWRRRAPCSTPRRPLTAAPPSA----CPSPASSPPPSRSWTPPTTPRAAPAGPLART 302
+P+P +R+ S RR + +PP A PSPA PPP PP R +P P R
Sbjct: 579 SPSPRKRQKETSPRRRRRSPSPPPARRRRSPSPAPPPPP----PPPPRRRRSPTPP-PRR 633
Query: 303 ACPSCPSTRCSSCWAWAA*SWRRRRT---PPC*ASSCPAC*AWRRATPA 440
PS P R S + +RR + PP ++ P RRA+P+
Sbjct: 634 RTPSPPPRRRSPSPRRYSPPIQRRYSPSPPPKRRTASPPPPPKRRASPS 682
Score = 37.7 bits (86), Expect = 0.22
Identities = 40/119 (33%), Positives = 51/119 (42%), Gaps = 11/119 (9%)
Frame = +3
Query: 3 TRPLTPSFTPSPA*TCCSACCWSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRR 182
TR +PS T + +SP P R P PPR P P R R S RR
Sbjct: 345 TRSRSPSHTRPRRRHRSRSRSYSPRRRPSPRRRPSPRRRTPPRRMPPPPRHRRSRSPGRR 404
Query: 183 ------PLTAAPPSACPSPASSP---PPSRSWTPP-TTPRAAP-AGPLARTACPSCPST 326
L+ + S+ S + SP PP R+ +PP T R +P A P R PS P+T
Sbjct: 405 RRRSSASLSGSSSSSSSSRSRSPPKKPPKRTSSPPRKTRRLSPSASPPRRRHRPSSPAT 463
Score = 37.4 bits (85), Expect = 0.29
Identities = 22/70 (31%), Positives = 33/70 (46%), Gaps = 5/70 (7%)
Frame = +3
Query: 135 TPTPWRRRAPCSTPRRPLTAAPPSACPSPASSPPPSRSWTPPTTP-----RAAPAGPLAR 299
+P P +R +P PR P T++PP ++SP +S +P T P R + +
Sbjct: 729 SPQPNKRHSPSPRPRAPQTSSPPPVRRGASASPQGRQSPSPSTRPIRRVSRTPEPKKIKK 788
Query: 300 TACPSCPSTR 329
A PS S R
Sbjct: 789 AASPSPQSVR 798
Score = 32.7 bits (73), Expect = 7.1
Identities = 28/108 (25%), Positives = 47/108 (42%), Gaps = 19/108 (17%)
Frame = -2
Query: 608 STHTRPAHNRTATSARRAV----------RSAQCPTRTSAKSRIRLAHQTANANT*FTRA 459
S RP+ R + RR R ++ P R +S L+ ++++++ +R+
Sbjct: 367 SPRRRPSPRRRPSPRRRTPPRRMPPPPRHRRSRSPGRRRRRSSASLSGSSSSSSSSRSRS 426
Query: 458 CPGSSQRRRSTPPRSARGAGRSSARGR--------APPPPGSRR-PRP 342
P +R S+PPR R S++ R A PPP +R P P
Sbjct: 427 PPKKPPKRTSSPPRKTRRLSPSASPPRRRHRPSSPATPPPKTRHSPTP 474
Score = 32.7 bits (73), Expect = 7.1
Identities = 35/109 (32%), Positives = 50/109 (45%), Gaps = 1/109 (0%)
Frame = -2
Query: 599 TRPAHNRTATSARRAVRSAQCPTRTSAKSRIRLAHQTANANT*FTRACPGSSQRRRSTPP 420
TRP +R+ + +R RS R+ + +R R H++ + + S RRR +P
Sbjct: 331 TRP-RSRSRSKSRSRTRS-----RSPSHTRPRRRHRSRSRSY---------SPRRRPSPR 375
Query: 419 RSARGAGRSSARGRAPPPPGSRRPR-PAR*APS*WAAWAGRSRQWSSRR 276
R R+ R R PPPP RR R P R A+ +G S SS R
Sbjct: 376 RRPSPRRRTPPR-RMPPPPRHRRSRSPGRRRRRSSASLSGSSSSSSSSR 423
Score = 32.7 bits (73), Expect = 7.1
Identities = 33/118 (27%), Positives = 45/118 (37%), Gaps = 28/118 (23%)
Frame = +3
Query: 66 WSPLAPPPAWRT**P*SCLPPRWTPTPWRRRAPCSTPRRPLTAAPPSACPSP-------- 221
+SP +PPP RT PP P P RR +P P+R ++ +PP SP
Sbjct: 658 YSP-SPPPKRRT-----ASPP---PPPKRRASPSPPPKRRVSHSPPPKQRSPPVTKRRSP 708
Query: 222 ------------------ASSPPPSRSWTPPTTPRA--APAGPLARTACPSCPSTRCS 335
A SP P++ +P PRA + P R + P R S
Sbjct: 709 SLSSKHRKGSSPGRSTREARSPQPNKRHSPSPRPRAPQTSSPPPVRRGASASPQGRQS 766