Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001272A_C01 KCC001272A_c01
(743 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
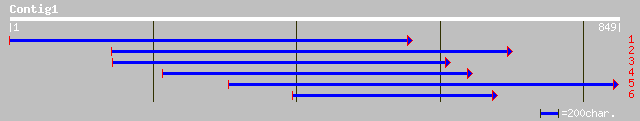
ref|ZP_00057775.1| COG1205: Distinct helicase family with a uniq... 47 3e-04
ref|NP_821742.1| hypothetical protein [Streptomyces avermitilis ... 45 0.001
emb|CAC17637.1| dJ1182A14.1 (similar to rat Espin) [Homo sapiens] 45 0.001
ref|NP_772549.1| blr5909 [Bradyrhizobium japonicum] gi|27354186|... 45 0.002
ref|NP_844716.1| conserved hypothetical protein [Bacillus anthra... 45 0.002
>ref|ZP_00057775.1| COG1205: Distinct helicase family with a unique C-terminal domain
including a metal-binding cysteine cluster [Thermobifida
fusca]
Length = 1958
Score = 47.0 bits (110), Expect = 3e-04
Identities = 63/203 (31%), Positives = 81/203 (39%), Gaps = 20/203 (9%)
Frame = +2
Query: 101 HSVRAS*AKGS*TAQVAAWDCCSRRKAVGGPQQDDQRQGIPEPVPCASGHRPILL----- 265
H R + A+ TA A +RR+A G ++ R RP+
Sbjct: 190 HRPRPAAARARRTAGGAGAQLRARRRATAGRRRRPARP-----------RRPLRRLRRTP 238
Query: 266 QPHPRRLHEGRHYGTHSR--GQQNRQQPHP--PGPV--SQGHCAH---VPRAPQPASDVQ 418
+PH R +GR G S G R+QP P P PV +Q H A +P +P +D +
Sbjct: 239 RPHARGSGQGRVAGRRSGAPGAAGRRQPLPGRPRPVRGAQQHRAAQGALPGGHRPEADRR 298
Query: 419 GHRELHRLLQRHRPGLLGQRWPNRLPHP------TRSGRSVRQCAHRCQPGR*GAGRLHL 580
G R R L G R P+R P P R R VR+ HR PGR G GRL
Sbjct: 299 GVRPAPRRLPDS-----GTR-PDRQPPPRPARVLARPARPVRRAGHRRPPGRPGRGRLPT 352
Query: 581 PAAVQVARVLGPAGKAVLARRWR 649
A V G + RR R
Sbjct: 353 AGGALPAGVGGTGRRGPRPRRRR 375
Score = 45.1 bits (105), Expect = 0.001
Identities = 46/146 (31%), Positives = 58/146 (39%), Gaps = 4/146 (2%)
Frame = +2
Query: 293 GRHYGTHSRGQQNRQQPHPPGPVSQGHCAHVPRAPQPASDVQGHRELHRLLQRHRPGLLG 472
GR RG + R++ P + H RA +P G R R RPG
Sbjct: 102 GRPARLRRRGPRRRRRLCPRPRRTPRH-----RAARPRLRRTGPRRQRPHPHRQRPG--- 153
Query: 473 QRWPNRLPHPTRSGRSVRQCAHRCQPGR*GAGRLH----LPAAVQVARVLGPAGKAVLAR 640
+R P R H R G +V A R +PG H PAA + R G AG + AR
Sbjct: 154 RRTPARPGHRGRGGPAVAAAARRGRPGHQRPAAPHPHRPRPAAARARRTAGGAGAQLRAR 213
Query: 641 RWRAGSAARERLEAPPVTPCARMSST 718
R +A R R A P P R+ T
Sbjct: 214 --RRATAGRRRRPARPRRPLRRLRRT 237
>ref|NP_821742.1| hypothetical protein [Streptomyces avermitilis MA-4680]
gi|29604206|dbj|BAC68277.1| hypothetical protein
[Streptomyces avermitilis MA-4680]
Length = 481
Score = 45.4 bits (106), Expect = 0.001
Identities = 48/168 (28%), Positives = 65/168 (38%), Gaps = 3/168 (1%)
Frame = +1
Query: 169 PSQSCRRPPTRRPAARDT*TCSLCVRTSAHPTSTSS*KAP*RASLWHTFTWPTKPPTTPS 348
P + R PP R PAA TC L ++ T T+S + P S TW T P
Sbjct: 346 PRRHPRSPPWRSPAA----TCLLPPSSAWAATGTTSSRCPVPGSHSWWATWSATASTPPP 401
Query: 349 AWA-CFPRSLRPCPPCSSTRL*RTRAP*TSPPPSTPQTWATGAALAQPTSSSN*ERAICT 525
WA C P PC P ++ T P ++ TW T ++ P
Sbjct: 402 PWAGCGP----PCAPSPTS---------TCRPTNSSPTWTTSSSACPP-----------K 437
Query: 526 SMCTPLPTRAVRCRAPSPASSRAGGPCARP--CREGRAS*ALARRICC 663
S+ PLP R P+P + +G P P A+ + AR CC
Sbjct: 438 SLPKPLPK-----RTPNPQGT-SGPPACTPYTIPSPAAAPSPARATCC 479
>emb|CAC17637.1| dJ1182A14.1 (similar to rat Espin) [Homo sapiens]
Length = 1130
Score = 45.4 bits (106), Expect = 0.001
Identities = 40/112 (35%), Positives = 48/112 (42%), Gaps = 3/112 (2%)
Frame = +1
Query: 325 TKPPTTPSAWACFPRSLRPCPPCSSTRL*RTRAP*TSPPPSTPQTWATGAALAQPTSSSN 504
T PPT+ + W PR++R P S AP ++ P TW ALA P
Sbjct: 746 TAPPTSCAHWTRRPRAVRAASPRSPL------APRSASPSCRRTTWRP--ALASPAPPPP 797
Query: 505 *ERAICTSMCTP---LPTRAVRCRAPSPASSRAGGPCARPCREGRAS*ALAR 651
R S TP L R R R SP+S RAG P AR C R + AR
Sbjct: 798 TARWPTGSPWTPWARLRHRIARRRYLSPSSWRAGRPSARNCAASRTTSTCAR 849
>ref|NP_772549.1| blr5909 [Bradyrhizobium japonicum] gi|27354186|dbj|BAC51174.1|
blr5909 [Bradyrhizobium japonicum USDA 110]
Length = 139
Score = 44.7 bits (104), Expect = 0.002
Identities = 25/45 (55%), Positives = 28/45 (61%), Gaps = 1/45 (2%)
Frame = +3
Query: 447 NATDLGYWGSAGPTDF-LIQLGAGDLYVNVHTVANPGGEVQGAFT 578
NAT GSA TD L AG LYVN+HT ANPGGE++G T
Sbjct: 94 NATSSPVEGSATLTDAQAADLLAGKLYVNIHTAANPGGEIRGQVT 138
>ref|NP_844716.1| conserved hypothetical protein [Bacillus anthracis str. Ames]
gi|30256970|gb|AAP26202.1| conserved hypothetical
protein [Bacillus anthracis str. Ames]
Length = 132
Score = 44.7 bits (104), Expect = 0.002
Identities = 29/96 (30%), Positives = 46/96 (47%), Gaps = 2/96 (2%)
Frame = +3
Query: 288 MKGVTMAHIHVANKTANNPIRLGLFPKVTAPMSPVLLNPPLTYKGTVNFTASFNATDLGY 467
++ V AH+H+ K N P+ LF +T P+S T DL
Sbjct: 43 IEDVVAAHLHIGAKGTNGPVVAFLFGPITNPVSIEC----------ATLTGMITQEDL-V 91
Query: 468 WGSAGPT--DFLIQLGAGDLYVNVHTVANPGGEVQG 569
AG T + ++ +G++Y+NVHTV +P GE++G
Sbjct: 92 GPLAGQTLGTLINEIISGNIYINVHTVQHPNGEIRG 127