Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001247A_C01 KCC001247A_c01
(608 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
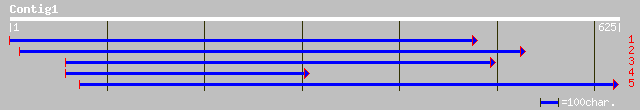
dbj|BAB32536.1| alpha-amylase [Drosophila melanogaster] 39 0.044
ref|NP_294953.1| hypothetical protein [Deinococcus radiodurans] ... 36 0.38
gb|AAK93556.1| SD08128p [Drosophila melanogaster] 36 0.38
ref|NP_649254.2| CG11399-PB [Drosophila melanogaster] gi|7296349... 36 0.38
gb|AAL25435.1| LD30158p [Drosophila melanogaster] 36 0.38
>dbj|BAB32536.1| alpha-amylase [Drosophila melanogaster]
Length = 195
Score = 39.3 bits (90), Expect = 0.044
Identities = 30/86 (34%), Positives = 40/86 (45%), Gaps = 1/86 (1%)
Frame = +3
Query: 219 SWAAWPRQRLSLMA-SSSSRPRSCLSGTSWLPRSCTWWPSPPLASLVSSAAATRFSTISW 395
SW P R S A SS++ P C +W S TW P+ +L ++ A I
Sbjct: 87 SWRPAPETRSSSPAWSSAATPSECAPTWTW--SSTTWPPTEAPTALAAAPPAPAARAIPE 144
Query: 396 R*THSC*SIVPRFASTRPSPSRTTTS 473
T PR+ STRP+PS TTT+
Sbjct: 145 CPT-------PRWTSTRPAPSATTTT 163
>ref|NP_294953.1| hypothetical protein [Deinococcus radiodurans]
gi|7472709|pir||F75420 hypothetical protein -
Deinococcus radiodurans (strain R1)
gi|6458982|gb|AAF10810.1|AE001971_10 hypothetical
protein [Deinococcus radiodurans]
Length = 319
Score = 36.2 bits (82), Expect = 0.38
Identities = 29/100 (29%), Positives = 38/100 (38%), Gaps = 9/100 (9%)
Frame = +3
Query: 219 SWAAWPRQRLSLMASSSSRPRSCLSGTSWLPRSCTWW---PSPPLASLVSSAAATRFSTI 389
S WP + + S PRS +GT W CTW P+ P A+ ++ + S
Sbjct: 219 STRCWPAASANSVPKPPSSPRSVRAGTRW--GCCTWTPGAPAAPRATTTRNSCSHSPSRR 276
Query: 390 SWR*T------HSC*SIVPRFASTRPSPSRTTTSTCQTGC 491
WR T S P RP+P TC T C
Sbjct: 277 VWRLTTPGSTPSRPASARPPRRCARPAPCWRAACTCPTRC 316
>gb|AAK93556.1| SD08128p [Drosophila melanogaster]
Length = 920
Score = 36.2 bits (82), Expect = 0.38
Identities = 37/139 (26%), Positives = 56/139 (39%), Gaps = 8/139 (5%)
Frame = +3
Query: 201 WRRGECSWAAWPRQRLSLMASSSS--------RPRSCLSGTSWLPRSCTWWPSPPLASLV 356
W + +A W + + A + R R+ + + S PS P S
Sbjct: 712 WLQNSAGYARWGPNEIRVEALREAFRPQRDRERERAAAAAAAAGVTSSQQSPSTP-PSAA 770
Query: 357 SSAAATRFSTISWR*THSC*SIVPRFASTRPSPSRTTTSTCQTGCTWHTSVHLLPGRFPA 536
S+A T TIS ++S S ST SPS TTTS+ + + HT P +FP
Sbjct: 771 STAPTTNSCTISGVNSNS--SSTATTTSTSSSPSLTTTSSSMSPLSPHTPTGNPPPQFPM 828
Query: 537 PLGTYS*LSSWALLAPRLS 593
+ S S+ +P +S
Sbjct: 829 TISNTSSSSNLVAASPTMS 847
>ref|NP_649254.2| CG11399-PB [Drosophila melanogaster] gi|7296349|gb|AAF51638.1|
CG11399-PB [Drosophila melanogaster]
Length = 920
Score = 36.2 bits (82), Expect = 0.38
Identities = 37/139 (26%), Positives = 56/139 (39%), Gaps = 8/139 (5%)
Frame = +3
Query: 201 WRRGECSWAAWPRQRLSLMASSSS--------RPRSCLSGTSWLPRSCTWWPSPPLASLV 356
W + +A W + + A + R R+ + + S PS P S
Sbjct: 712 WLQNSAGYARWGPNEIRVEALREAFRPQRDRERERAAAAAAAAGVTSSQQSPSTP-PSAA 770
Query: 357 SSAAATRFSTISWR*THSC*SIVPRFASTRPSPSRTTTSTCQTGCTWHTSVHLLPGRFPA 536
S+A T TIS ++S S ST SPS TTTS+ + + HT P +FP
Sbjct: 771 STAPTTNSCTISGVNSNS--SSTATTTSTSSSPSLTTTSSSMSPLSPHTPTGNPPPQFPM 828
Query: 537 PLGTYS*LSSWALLAPRLS 593
+ S S+ +P +S
Sbjct: 829 TISNTSSSSNLVAASPTMS 847
>gb|AAL25435.1| LD30158p [Drosophila melanogaster]
Length = 243
Score = 36.2 bits (82), Expect = 0.38
Identities = 37/139 (26%), Positives = 56/139 (39%), Gaps = 8/139 (5%)
Frame = +3
Query: 201 WRRGECSWAAWPRQRLSLMASSSS--------RPRSCLSGTSWLPRSCTWWPSPPLASLV 356
W + +A W + + A + R R+ + + S PS P S
Sbjct: 35 WLQNSAGYARWGPNEIRVEALREAFRPQRDRERERAAAAAAAAGVTSSQQSPSTP-PSAA 93
Query: 357 SSAAATRFSTISWR*THSC*SIVPRFASTRPSPSRTTTSTCQTGCTWHTSVHLLPGRFPA 536
S+A T TIS ++S S ST SPS TTTS+ + + HT P +FP
Sbjct: 94 STAPTTNSCTISGVNSNS--SSTATTTSTSSSPSLTTTSSSMSPLSPHTPTGNPPPQFPM 151
Query: 537 PLGTYS*LSSWALLAPRLS 593
+ S S+ +P +S
Sbjct: 152 TISNTSSSSNLVAASPTMS 170