Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001237A_C01 KCC001237A_c01
(1097 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
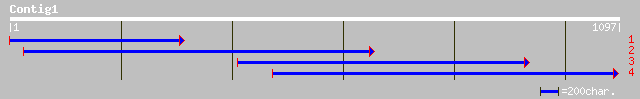
dbj|BAC86069.1| unnamed protein product [Homo sapiens] 47 6e-04
gb|EAA39492.1| GLP_26_54603_52153 [Giardia lamblia ATCC 50803] 45 0.002
ref|XP_308046.1| ENSANGP00000019335 [Anopheles gambiae] gi|21291... 44 0.007
ref|ZP_00091362.1| COG2192: Predicted carbamoyl transferase, Nod... 43 0.008
ref|ZP_00058502.1| hypothetical protein [Thermobifida fusca] 42 0.014
>dbj|BAC86069.1| unnamed protein product [Homo sapiens]
Length = 201
Score = 47.0 bits (110), Expect = 6e-04
Identities = 52/164 (31%), Positives = 63/164 (37%), Gaps = 21/164 (12%)
Frame = +2
Query: 515 HRGRRWLGLAGPRVQLHS-------AAL*GSGDVAATRHRGRRRGRGE*RRLRGA---GV 664
HRG WLG G S A G G V A + R RR G R GA G
Sbjct: 19 HRGATWLGRGGGAGPGQSGPGHGQGTATPGPGQVRAAQARARRGGGARERPAEGAREPGA 78
Query: 665 ARQP*RAVPTTLNQLDAPYMIDAPEDCP-----LTSSAYSFSSDRQTPRRGAHGQPVQPL 829
AR+ RA P L A + A P + F RQ+PR +HGQP + L
Sbjct: 79 ARRGARAGPGA--HLRAARTLRARAQTPQLRREAACAGSFFPKCRQSPR--SHGQPFRQL 134
Query: 830 HPPRRVER-GW-----RGQRPQRARQLRAASSAGPPPNPAQAGR 943
+ R GW G+RP R + A + G P GR
Sbjct: 135 QSHWQAPRQGWPAQEGGGRRPSRREEAAALAGGGKRTAPGLRGR 178
>gb|EAA39492.1| GLP_26_54603_52153 [Giardia lamblia ATCC 50803]
Length = 816
Score = 45.4 bits (106), Expect = 0.002
Identities = 59/174 (33%), Positives = 67/174 (37%), Gaps = 1/174 (0%)
Frame = -3
Query: 690 GTARQGCRATPAPLSRRHSPRPRRR-PLCLVAATSPDPQRAAECS*TRGPARPSHRRPRW 514
G R R P P RR PRPRRR PL +PD +R RGP R PR
Sbjct: 309 GPPRGDARRGPPPRPRR--PRPRRRAPLRSARRGAPDARRRRRRV-CRGPPRGRGGAPR- 364
Query: 513 RAGCATRLGAPPI*TSAPARPW*AAYICCSQE*QPLAGPGQRQEQALSRRRRSAPRRPCR 334
G A APP ARP P G + +A + R R A R P R
Sbjct: 365 --GPARLPRAPP----PAARPG-----------PPPPGRALVRARAPAARLRGARRGPPR 407
Query: 333 RLLPRFPGAELHLPGSSRSQDCC*PAAPGCGPCCAAR*AQSRDRPGRRREERSG 172
L R P A +P R+ P AP P + RPGRRR R G
Sbjct: 408 GALRRRPRARPRVPRVLRAPRRDRPVAPRPRPVRRGP-PGAEARPGRRRGRRGG 460
Score = 34.3 bits (77), Expect = 3.9
Identities = 81/288 (28%), Positives = 101/288 (34%), Gaps = 36/288 (12%)
Frame = +2
Query: 191 LRPGRSLLCAQRAAQQGPQPG-AAGQQQSWLLLLPGRCSSAPGKR---------GSRRRH 340
+ P R+ LC R P+PG G Q++ P PG+R G
Sbjct: 1 MHPARARLCDVRRRPGAPRPGRPRGAQRA---PRPAPDPPGPGRRPPRAPGALGGMLPAA 57
Query: 341 GRL-GADRRRRLRACSCRWPGPARGCHSC--EQQMYAAHHGRAGADV*IGGAPKRVAHPA 511
GRL G R R LRA S G RG + A GR G +G R P
Sbjct: 58 GRLAGPARGRGLRAAS----GALRGVRPVLRPRGRPVARPGRVGRGGGLGRL--RDGRPR 111
Query: 512 RHRGRRWLGLAGPRVQLHSAAL*GSGDVAATRHRGRRRGRGE*RRLRGAGVARQP*RAVP 691
+ R LG G R GD R R RRG G R A V R+P RA
Sbjct: 112 QRAPGRALGAPGRRRPPAGRRAPVRGDRRRGRPRAPRRGPGG-RLGPRAPVCRRPRRAGR 170
Query: 692 TTLNQLDAPYMIDA-------PEDCPLTSSAYSFSSDRQTPR--RGAHGQP--------- 817
+ AP + P P +R PR G HG P
Sbjct: 171 GAPGR--APGRVPVRRRVPARPAPPPAARYRVRALPERGAPRGAPGRHGPPGLRPRGRGA 228
Query: 818 ---VQPLHPPRR--VERGWRGQRPQRARQLRAASSAGPPPNPAQAGRS 946
+PL PR RG R + + A + R ++ GPP A GR+
Sbjct: 229 CARRRPLPVPRARGPPRGVRARLGRDAAR-RGVAARGPPARAAAPGRA 275
Score = 33.9 bits (76), Expect = 5.2
Identities = 75/270 (27%), Positives = 95/270 (34%), Gaps = 23/270 (8%)
Frame = +2
Query: 197 PGRSLLCAQRAAQQGPQPGAAGQQQSWLLLLPGRCSSAPGKRGSRRRHGRLGADRRRRLR 376
PGR+ R + GP+P + GR S PG+RG R R G R R
Sbjct: 272 PGRA---GDRRGRPGPRPRVRAPLRP--RHADGRLSPPPGRRGPPRGDARRGPPPRPRRP 326
Query: 377 ACSCRWP--GPARGCHSCEQQMYAAHHGRAGADV*IGGAPK------RVAHPARHRGRRW 532
R P RG ++ G GGAP+ R PA G
Sbjct: 327 RPRRRAPLRSARRGAPDARRRRRRVCRGPPRGR---GGAPRGPARLPRAPPPAARPGPPP 383
Query: 533 LGLAGPRVQLHSAAL*GSGDVAATRHRGRRRGRGE*RRLRGAGVARQP*RAVPTTLNQLD 712
G A R + A R RG RRG RGA + R+P RA P L
Sbjct: 384 PGRALVRAR-----------APAARLRGARRGPP-----RGA-LRRRP-RARPRVPRVLR 425
Query: 713 APYMID--APEDCPLTSSAYSFSSDRQTPRRGAHGQP-------VQPLHPPRRVER---- 853
AP AP P+ + R RRG G P Q P R R
Sbjct: 426 APRRDRPVAPRPRPVRRGPPG-AEARPGRRRGRRGGPPTSGLAFTQSGAPDPRAARRPAT 484
Query: 854 --GWRGQRPQRARQLRAASSAGPPPNPAQA 937
R RA + RA ++AG PP+ +++
Sbjct: 485 SPRSRATSSSRALKRRAVATAGAPPSASRS 514
>ref|XP_308046.1| ENSANGP00000019335 [Anopheles gambiae] gi|21291511|gb|EAA03656.1|
ENSANGP00000019335 [Anopheles gambiae str. PEST]
Length = 414
Score = 43.5 bits (101), Expect = 0.007
Identities = 66/273 (24%), Positives = 99/273 (36%), Gaps = 23/273 (8%)
Frame = +1
Query: 172 ARTFFAPATRPISTLCSARCTARTATRSCRSTTILASTTSWQMQFSPREARQ-----QAS 336
ART A AT T SA T + A+ S + T T+ +P+ + + +
Sbjct: 123 ARTKAASATSTTKTATSASSTTKAASASSVTRTAAPKPTTGSAGAAPKPSSRLSTASSTT 182
Query: 337 TRASRCRPAAAAQSLFLPLARSGERLSFL*AANVRCSPRTCWRRCLNWRRTQTCRTSCSP 516
TR RP++A S +P S + S T + T+T +T+ S
Sbjct: 183 TRTVTSRPSSAVSSASVPSKTSTTTTTTAKRTVAAKSSLTNGTSATDGAATRTTKTTTSS 242
Query: 517 SWT-TMARPRWSAC------------STTFS--RPLRVRRRCRH*TQRAAPWAGRVTTTE 651
+ T A R SA STT + +PL + T ++ AGR TTT
Sbjct: 243 TVPRTGAATRTSATAGSGLAGKTSSSSTTAAAKKPLLDAKTTASKTASSSTAAGRTTTTT 302
Query: 652 GGGSGAAALTSGTNHFKPA*RTIHDRRARGLPAHELCIQLQ*---RPADAKTGRTWAACA 822
A T+G P+ R + + + +PA + A A
Sbjct: 303 RTSLAPKAATAGAT--SPSVRKVQQNGVAAKKSTTSAVSPAPGTKKPAPITRPKGTATTA 360
Query: 823 AATPPAACRAGLAGPTTPAGSPTSRCELGRPAP 921
AA P AA G+ G T A P + +P
Sbjct: 361 AAAPAAAKTNGVVGTVTEAAVPVDAAPVAVTSP 393
Score = 33.9 bits (76), Expect = 5.2
Identities = 58/261 (22%), Positives = 91/261 (34%), Gaps = 9/261 (3%)
Frame = +1
Query: 187 APATRPISTLCSARCTARTATRSCRSTTILASTTSWQMQFSPREARQQASTRASRCRPAA 366
APAT+ T S+ ART S STT A++ S + + A + TR + +P
Sbjct: 110 APATK---TTASSVAAARTKAASATSTTKTATSAS----STTKAASASSVTRTAAPKPTT 162
Query: 367 AAQSLFLPLARSGERLSFL*AANVRCSPRTCWRRCLNWRRTQTCRTSCSPSWTTMARPRW 546
+ + RLS + RT R + + + + S + TT A+
Sbjct: 163 GSAG---AAPKPSSRLS----TASSTTTRTVTSRPSSAVSSASVPSKTSTTTTTTAKRTV 215
Query: 547 SACST----TFSRPLRVRRRCRH*TQRAAPWAGRVTTTEG-GGSGAAALTSGTNHFKPA* 711
+A S+ T + R + T P G T T GSG A TS ++ A
Sbjct: 216 AAKSSLTNGTSATDGAATRTTKTTTSSTVPRTGAATRTSATAGSGLAGKTSSSSTTAAAK 275
Query: 712 RTIHDRRARGLPAHELCIQLQ*RPADAKTGRTWAACAAATPPAACRAGLAGPTT----PA 879
+ + D + + GRT + P A AG P+
Sbjct: 276 KPLLDAKTTASKT---------ASSSTAAGRTTTTTRTSLAPKAATAGATSPSVRKVQQN 326
Query: 880 GSPTSRCELGRPAPKPSASRP 942
G + +P P +P
Sbjct: 327 GVAAKKSTTSAVSPAPGTKKP 347
>ref|ZP_00091362.1| COG2192: Predicted carbamoyl transferase, NodU family [Azotobacter
vinelandii]
Length = 1081
Score = 43.1 bits (100), Expect = 0.008
Identities = 82/298 (27%), Positives = 103/298 (34%), Gaps = 41/298 (13%)
Frame = +2
Query: 170 RPERSSRLRPGR-------SLLCAQRAAQQGPQPGAAGQQQSWLLLLPGRCS-------- 304
R R+ RL G+ + LC R + G A ++ + P R
Sbjct: 29 RRRRTGRLASGQPAARAAGTTLCRSRRVARPAGAGQAFGRRQGVAAFPARAEGCAPARRP 88
Query: 305 ----SAPGKRGSRRRHGRLGADRRRRLRACSCRWPGPARGCHSCEQQMYAAHHGRAGADV 472
+AP +RG+ R GRL A R R R PGP RG A GRA A
Sbjct: 89 AAGHAAPARRGAGRGRGRLAAVRVSR------RGPGPERG--------LATGGGRAAAVR 134
Query: 473 *IGGAPKRVAHPARHRGRRWLGLAGPRVQLHSAAL*GSGDVAATRHRGRRR-GRGE*RR- 646
G +R + R RR LAG AA G RGRRR G G R
Sbjct: 135 RPAGGTRRGSGRHRRPARRGT-LAGRHAPGQPAAPRGPA------VRGRRRCGAGTDARQ 187
Query: 647 ----LRGAGVARQP*RAVPTTLNQLDAPYMIDAPEDCPLTSSAYSFSSDRQTP------R 796
GAG R+ R VP L T+ A R P +
Sbjct: 188 AAVARAGAGEPRRLLRPVPGRARALHRG-----------TAGALPAGQRRARPAAGGAAQ 236
Query: 797 RGAHGQPVQP----------LHPPRRVERGWRGQRPQRARQLRAASSAGPPPNPAQAG 940
G G P+ P LHP +R R R ++ AA +AG P +AG
Sbjct: 237 GGCQGAPLAPARLPRQARPRLHPVQRAARAHGAARGAPRQRGPAAPAAGRPGCAHRAG 294
>ref|ZP_00058502.1| hypothetical protein [Thermobifida fusca]
Length = 1903
Score = 42.4 bits (98), Expect = 0.014
Identities = 51/175 (29%), Positives = 59/175 (33%), Gaps = 6/175 (3%)
Frame = -3
Query: 651 LSRRHSPRPRRRPLCLVAATSPDPQRAA--ECS*TRGPARPSHRRPRWRAGCATRLGAPP 478
++R H R +RRP A P P R R P RP R PR R G T P
Sbjct: 283 VARPHHHRAQRRPRPRAAPAHPAPHRPRLPRDRLPRPPRRPETRHPRRRRGRRT-----P 337
Query: 477 I*TSAPARPW*AAYICCSQE*QPLAGPGQRQEQALSRRRRSAPRRPCRRLLPRFPGAELH 298
+ PARP G R + + PR P RR L R P
Sbjct: 338 VPLPRPARP----------------HRGTRPRKRAAAGTHRPPRPPRRRPLARLPARSGK 381
Query: 297 LPGSSRSQDCC*PAAPGCGPCCAAR*AQSRDRPGRRREE----RSGRSEASGATK 145
G RS P P R RDR R R R GR+ GA +
Sbjct: 382 ERGPRRSH-------PARHPLRIRRLGAHRDRSRRARRAHGRLRRGRAPRLGAAR 429