Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001170A_C01 KCC001170A_c01
(518 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
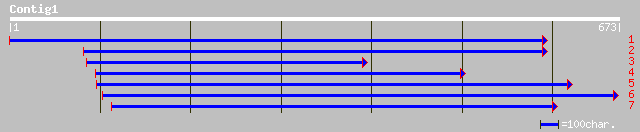
sp|P13728|SGS3_DROYA Salivary glue protein SGS-3 precursor gi|85... 45 4e-04
ref|XP_320719.1| ENSANGP00000020181 [Anopheles gambiae] gi|30174... 44 0.002
ref|NP_041080.1| membrane glycoprotein [Equine herpesvirus 1] gi... 43 0.002
gb|AAK61485.1| glycoprotein gp2 [Equine herpesvirus 1] 43 0.002
pir||T45462 membrane glycoprotein [imported] - equine herpesviru... 43 0.002
>sp|P13728|SGS3_DROYA Salivary glue protein SGS-3 precursor gi|85255|pir||S01360 salivary
glue protein sgs-3 precursor - fruit fly (Drosophila
yakuba)
Length = 263
Score = 45.4 bits (106), Expect = 4e-04
Identities = 42/144 (29%), Positives = 63/144 (43%), Gaps = 17/144 (11%)
Frame = +3
Query: 78 QSTTRRALS*TR*TPS*ASTLSALTTTSQTPRTRCSDCER--------RARARKRTCSRR 233
+STTRR + TR T + +T + TTT++ P TR + R T +RR
Sbjct: 79 KSTTRRTTTTTRQTTTRPTTTTTTTTTTRRPTTRSTTTRHTTTTTTTTRRPTTTTTTTRR 138
Query: 234 ---CCSSSRRPAVLT------RTRSRSYQRLWSSCASATPTHTLTTRHTSLARSRSATLT 386
+++RRP T TRS + + S S PTH TT TS ++ T T
Sbjct: 139 PTTTTTTTRRPTTTTTTTRLPTTRSTTTRHTTKSTTSKRPTHETTT--TSKRPTQETTTT 196
Query: 387 CRPSATCSR**SSQPRPPSPPRAG 458
R + ++ P+P + P G
Sbjct: 197 TRRATQA----TTTPKPTNKPGCG 216
Score = 38.5 bits (88), Expect = 0.052
Identities = 31/105 (29%), Positives = 50/105 (47%), Gaps = 10/105 (9%)
Frame = +3
Query: 120 PS*ASTLSALTTTSQTPRTR-----CSDCERRA-RARKRTCSRRCCSSSR----RPAVLT 269
P+ A T + TTT+ P TR C+D R +++ +RR +++R RP T
Sbjct: 41 PTCAPTTTTTTTTTCAPPTRPPPPPCTDAPTTTKRTTEKSTTRRTTTTTRQTTTRPTTTT 100
Query: 270 RTRSRSYQRLWSSCASATPTHTLTTRHTSLARSRSATLTCRPSAT 404
T + + + + S T HT TT T+ + + T T RP+ T
Sbjct: 101 TTTTTTRR---PTTRSTTTRHTTTTTTTTRRPTTTTTTTRRPTTT 142
>ref|XP_320719.1| ENSANGP00000020181 [Anopheles gambiae] gi|30174160|gb|EAA00415.2|
ENSANGP00000020181 [Anopheles gambiae str. PEST]
Length = 340
Score = 43.5 bits (101), Expect = 0.002
Identities = 52/167 (31%), Positives = 67/167 (39%), Gaps = 10/167 (5%)
Frame = -1
Query: 518 ARATATVPKTCVAWPDLPLSARPRWARRAGLA*LSARARCRRTTGQSCTSRSR*RGMS-- 345
AR T P C + AR RW+ + A ++RARCR + + +R R S
Sbjct: 21 ARRTRCAPSACRSCR----CARSRWSSPSS-ARTTSRARCRPSRTATGRARRSLRSSSST 75
Query: 344 --RCKRVRRRCARA*AP*ALVAPRTRPCQHRGTPRTAAAAP*ARPLSCPCPSFAVATPRA 171
R RR C R +P A A +HR T R A R + P P TPR
Sbjct: 76 AVRSTSGRRCCGRPRSPSATRAT----LRHRRTRRRAV-----RGMRRPRPP----TPRT 122
Query: 170 RRLRGGCKGRKGACLGRRSTRS------RQSPPCCRLRPPRTT*PCS 48
R GC+ R R TRS R++ CR RP R P +
Sbjct: 123 RTTTAGCRVRPSFPSSRSRTRSTCTTVRRRTRALCRERPDRPDRPAT 169
Score = 36.2 bits (82), Expect = 0.26
Identities = 36/104 (34%), Positives = 45/104 (42%)
Frame = +3
Query: 144 ALTTTSQTPRTRCSDCERRARARKRTCSRRCCSSSRRPAVLTRTRSRSYQRLWSSCASAT 323
A S +PRT S ARAR+ C+ C S R +R S S R +S A
Sbjct: 3 ASRAASTSPRTASS---TTARARRTRCAPSACRSCR--CARSRWSSPSSART-TSRARCR 56
Query: 324 PTHTLTTRHTSLARSRSATLTCRPSATCSR**SSQPRPPSPPRA 455
P+ T T R RS S+T +T R +PR PS RA
Sbjct: 57 PSRTATGRARRSLRSSSSTAV---RSTSGRRCCGRPRSPSATRA 97
>ref|NP_041080.1| membrane glycoprotein [Equine herpesvirus 1]
gi|138350|sp|P28968|VGLX_HSVEB GLYCOPROTEIN X PRECURSOR
gi|73738|pir||VGBEX1 glycoprotein X precursor - equine
herpesvirus 1 (strain Ab4p) gi|330862|gb|AAB02506.1|
membrane glycoprotein
Length = 797
Score = 43.1 bits (100), Expect = 0.002
Identities = 34/116 (29%), Positives = 50/116 (42%)
Frame = +3
Query: 57 SSCARWTQSTTRRALS*TR*TPS*ASTLSALTTTSQTPRTRCSDCERRARARKRTCSRRC 236
S+ ++T A + T TP+ ST +A TT++ TP + + E A T +
Sbjct: 267 STSTTGASTSTPSASTATSATPTSTSTSAAATTSTPTPTSAATSAESTTEAPTSTPTTDT 326
Query: 237 CSSSRRPAVLTRTRSRSYQRLWSSCASATPTHTLTTRHTSLARSRSATLTCRPSAT 404
+ S T S + +S SAT T T HTS S +T T PS+T
Sbjct: 327 TTPSEATTATTSPESTTVS---ASTTSATTTAFTTESHTSPDSSTGSTSTAEPSST 379
>gb|AAK61485.1| glycoprotein gp2 [Equine herpesvirus 1]
Length = 826
Score = 43.1 bits (100), Expect = 0.002
Identities = 34/116 (29%), Positives = 50/116 (42%)
Frame = +3
Query: 57 SSCARWTQSTTRRALS*TR*TPS*ASTLSALTTTSQTPRTRCSDCERRARARKRTCSRRC 236
S+ ++T A + T TP+ ST +A TT++ TP + + E A T +
Sbjct: 296 STSTTGASTSTPSASTATSATPTSTSTSAAATTSTPTPTSAATSAESTTEAPTSTPTTDT 355
Query: 237 CSSSRRPAVLTRTRSRSYQRLWSSCASATPTHTLTTRHTSLARSRSATLTCRPSAT 404
+ S T S + +S SAT T T HTS S +T T PS+T
Sbjct: 356 TTPSEATTATTSPESTTVS---ASTTSATTTAFTTESHTSPDSSTGSTSTAEPSST 408
>pir||T45462 membrane glycoprotein [imported] - equine herpesvirus 1
gi|2114321|dbj|BAA20037.1| membrane glycoprotein [Equine
herpesvirus 1]
Length = 866
Score = 43.1 bits (100), Expect = 0.002
Identities = 34/116 (29%), Positives = 50/116 (42%)
Frame = +3
Query: 57 SSCARWTQSTTRRALS*TR*TPS*ASTLSALTTTSQTPRTRCSDCERRARARKRTCSRRC 236
S+ ++T A + T TP+ ST +A TT++ TP + + E A T +
Sbjct: 336 STSTTGASTSTPSASTATSATPTSTSTSAAATTSTPTPTSAATSAESTTEAPTSTPTTDT 395
Query: 237 CSSSRRPAVLTRTRSRSYQRLWSSCASATPTHTLTTRHTSLARSRSATLTCRPSAT 404
+ S T S + +S SAT T T HTS S +T T PS+T
Sbjct: 396 TTPSEATTATTSPESTTVS---ASTTSATTTAFTTESHTSPDSSTGSTSTAEPSST 448