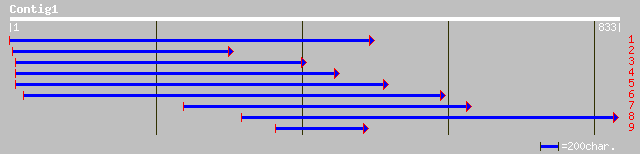
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001145A_C01 KCC001145A_c01
(832 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAN71768.1| ClpP2 [Synechococcus sp. PCC 7942] 238 1e-61
sp|O34125|CLP2_SYNP7 ATP-dependent Clp protease proteolytic subu... 236 3e-61
dbj|BAA85451.1| S-locus protein 2 [Brassica rapa] 229 5e-59
emb|CAB89185.1| ClpP [Brassica napus var. napus] 228 9e-59
emb|CAC80640.1| ClpP putative protein [Brassica napus] 228 9e-59
>gb|AAN71768.1| ClpP2 [Synechococcus sp. PCC 7942]
Length = 302
Score = 238 bits (606), Expect = 1e-61
Identities = 131/265 (49%), Positives = 169/265 (63%), Gaps = 11/265 (4%)
Frame = +3
Query: 63 RPRPHPQFPSF*VTRSLPMVAAALLGGLQGACPLVAGKRSARRACVPKAQLHDSQRPKGW 242
R +P P + S P+ ++G + PL+ S+ + +H + G
Sbjct: 28 RKKPKP------LAHSRPVRRGIIVGSGTLSLPLLYPVPSSMFSSHASLPIHSRRLAAGL 81
Query: 243 DRPWIQANSQPI-----VAPRTAEMQG------DPFGLLLRQRIVFLGGEVEDFGADAII 389
D W Q Q I + P E G D + LLR+RIVFLG V+D AD+I+
Sbjct: 82 DSRWQQPQIQAIAGSQAIVPTVVEQSGRGERAFDIYSRLLRERIVFLGTGVDDAVADSIV 141
Query: 390 SQLLLLDSQDPTKDIKIFINSPGGSVTAGMGIYDAMMLCRADVNTYCFGLAASMGAFLLG 569
+QLL L+++DP KDI+++INSPGGSVTAGM IYD M DV T CFGLAASMGAFLL
Sbjct: 142 AQLLFLEAEDPEKDIQLYINSPGGSVTAGMAIYDTMQQVAPDVATICFGLAASMGAFLLS 201
Query: 570 AGKRGKRNSMPNSRIMIHQPLGGASGQAVDIEIQAKEIMYHKANLNRIMADYCQQPLSKI 749
G +GKR ++P++RIMIHQPLGGA GQAVDIEIQA+EI+YHK+ LN ++A + QPL KI
Sbjct: 202 GGAQGKRMALPSARIMIHQPLGGAQGQAVDIEIQAREILYHKSTLNDLLAQHTGQPLEKI 261
Query: 750 EEDTDRDRYMSPLEAKEYGLIDHII 824
E DTDRD +MSP EAK YGLID ++
Sbjct: 262 EVDTDRDFFMSPEEAKAYGLIDQVL 286
>sp|O34125|CLP2_SYNP7 ATP-dependent Clp protease proteolytic subunit 2 (Endopeptidase Clp
2) gi|2351823|gb|AAB68677.1| ATP-dependent Clp protease,
proteolytic subunit [Synechococcus sp. PCC 7942]
gi|15620530|gb|AAL03914.1|U30252_2 ClpP2 [Synechococcus
sp. PCC 7942]
Length = 240
Score = 236 bits (602), Expect = 3e-61
Identities = 124/203 (61%), Positives = 151/203 (74%), Gaps = 8/203 (3%)
Frame = +3
Query: 240 WDRPWIQA--NSQPIVAPRTAEMQG------DPFGLLLRQRIVFLGGEVEDFGADAIISQ 395
W +P IQA SQ IV P E G D + LLR+RIVFLG V+D AD+I++Q
Sbjct: 23 WQQPQIQAIAGSQAIV-PTVVEQSGRGERAFDIYSRLLRERIVFLGTGVDDAVADSIVAQ 81
Query: 396 LLLLDSQDPTKDIKIFINSPGGSVTAGMGIYDAMMLCRADVNTYCFGLAASMGAFLLGAG 575
LL L+++DP KDI+++INSPGGSVTAGM IYD M DV T CFGLAASMGAFLL G
Sbjct: 82 LLFLEAEDPEKDIQLYINSPGGSVTAGMAIYDTMQQVAPDVATICFGLAASMGAFLLSGG 141
Query: 576 KRGKRNSMPNSRIMIHQPLGGASGQAVDIEIQAKEIMYHKANLNRIMADYCQQPLSKIEE 755
+GKR ++P++RIMIHQPLGGA GQAVDIEIQA+EI+YHK+ LN ++A + QPL KIE
Sbjct: 142 AQGKRMALPSARIMIHQPLGGAQGQAVDIEIQAREILYHKSTLNDLLAQHTGQPLEKIEV 201
Query: 756 DTDRDRYMSPLEAKEYGLIDHII 824
DTDRD +MSP EAK YGLID ++
Sbjct: 202 DTDRDFFMSPEEAKAYGLIDQVL 224
>dbj|BAA85451.1| S-locus protein 2 [Brassica rapa]
Length = 311
Score = 229 bits (583), Expect = 5e-59
Identities = 119/224 (53%), Positives = 154/224 (68%), Gaps = 8/224 (3%)
Frame = +3
Query: 177 RSARRACVPKAQLHDSQRPK-----GWDRPWIQANS---QPIVAPRTAEMQGDPFGLLLR 332
+ A+ CV + RP+ WD +S P P E+ D +LLR
Sbjct: 37 KRAKPFCVRSSMNLSGSRPRQTLSSNWDVSKFSVDSVAQSPSRLPSFEEL--DTTNMLLR 94
Query: 333 QRIVFLGGEVEDFGADAIISQLLLLDSQDPTKDIKIFINSPGGSVTAGMGIYDAMMLCRA 512
QRIVFLG +V+D AD +ISQLLLLD+QD +DI +FINSPGGS+TAGMGIYDAM C+A
Sbjct: 95 QRIVFLGSQVDDMTADLVISQLLLLDAQDSERDITLFINSPGGSITAGMGIYDAMKQCKA 154
Query: 513 DVNTYCFGLAASMGAFLLGAGKRGKRNSMPNSRIMIHQPLGGASGQAVDIEIQAKEIMYH 692
DV+T C GLAASMGAFLL +G +GKR MPNS++MIHQPLG A G+A ++ I+ +E+MYH
Sbjct: 155 DVSTVCLGLAASMGAFLLASGSKGKRYCMPNSKVMIHQPLGSAGGKATEMSIRVREMMYH 214
Query: 693 KANLNRIMADYCQQPLSKIEEDTDRDRYMSPLEAKEYGLIDHII 824
K LN+I + +P S+IE DTDRD +++P EAKEYGL+D +I
Sbjct: 215 KIKLNKIFSRITGKPESEIEGDTDRDYFLNPWEAKEYGLVDAVI 258
>emb|CAB89185.1| ClpP [Brassica napus var. napus]
Length = 313
Score = 228 bits (581), Expect = 9e-59
Identities = 119/224 (53%), Positives = 154/224 (68%), Gaps = 8/224 (3%)
Frame = +3
Query: 177 RSARRACVPKAQLHDSQRPK-----GWDRPWIQANS---QPIVAPRTAEMQGDPFGLLLR 332
+ A+ CV + RP+ WD +S P P E+ D +LLR
Sbjct: 39 KRAKPFCVRSSMNLSGPRPRQTLSSDWDVSKFSVDSVAQSPSRLPSFEEL--DTTNMLLR 96
Query: 333 QRIVFLGGEVEDFGADAIISQLLLLDSQDPTKDIKIFINSPGGSVTAGMGIYDAMMLCRA 512
QRIVFLG +V+D AD +ISQLLLLD+QD +DI +FINSPGGS+TAGMGIYDAM C+A
Sbjct: 97 QRIVFLGSQVDDMTADLVISQLLLLDAQDSERDITLFINSPGGSITAGMGIYDAMKQCKA 156
Query: 513 DVNTYCFGLAASMGAFLLGAGKRGKRNSMPNSRIMIHQPLGGASGQAVDIEIQAKEIMYH 692
DV+T C GLAASMGAFLL +G +GKR MPNS++MIHQPLG A G+A ++ I+ +E+MYH
Sbjct: 157 DVSTVCLGLAASMGAFLLASGSKGKRYCMPNSKVMIHQPLGSAGGKATEMSIRVREMMYH 216
Query: 693 KANLNRIMADYCQQPLSKIEEDTDRDRYMSPLEAKEYGLIDHII 824
K LN+I + +P S+IE DTDRD +++P EAKEYGL+D +I
Sbjct: 217 KIKLNKIFSRITGKPESEIEGDTDRDYFLNPWEAKEYGLVDAVI 260
>emb|CAC80640.1| ClpP putative protein [Brassica napus]
Length = 313
Score = 228 bits (581), Expect = 9e-59
Identities = 119/224 (53%), Positives = 154/224 (68%), Gaps = 8/224 (3%)
Frame = +3
Query: 177 RSARRACVPKAQLHDSQRPK-----GWDRPWIQANS---QPIVAPRTAEMQGDPFGLLLR 332
+ A+ CV + RP+ WD +S P P E+ D +LLR
Sbjct: 39 KRAKPFCVRSSMNLSGPRPRQTLSSDWDVSKFSVDSVAQSPSRLPSFEEL--DTTNMLLR 96
Query: 333 QRIVFLGGEVEDFGADAIISQLLLLDSQDPTKDIKIFINSPGGSVTAGMGIYDAMMLCRA 512
QRIVFLG +V+D AD +ISQLLLLD+QD +DI +FINSPGGS+TAGMGIYDAM C+A
Sbjct: 97 QRIVFLGSQVDDMTADLVISQLLLLDAQDSERDITLFINSPGGSITAGMGIYDAMKQCKA 156
Query: 513 DVNTYCFGLAASMGAFLLGAGKRGKRNSMPNSRIMIHQPLGGASGQAVDIEIQAKEIMYH 692
DV+T C GLAASMGAFLL +G +GKR MPNS++MIHQPLG A G+A ++ I+ +E+MYH
Sbjct: 157 DVSTVCLGLAASMGAFLLASGSKGKRYCMPNSKVMIHQPLGSAGGKATEMSIRVREMMYH 216
Query: 693 KANLNRIMADYCQQPLSKIEEDTDRDRYMSPLEAKEYGLIDHII 824
K LN+I + +P S+IE DTDRD +++P EAKEYGL+D +I
Sbjct: 217 KIKLNKIFSRITGKPESEIEGDTDRDYFLNPWEAKEYGLVDAVI 260