Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001097A_C01 KCC001097A_c01
(995 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
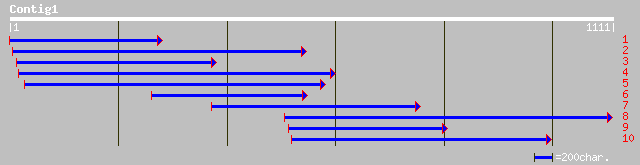
ref|NP_197703.1| photosystem II stability/assembly factor HCF136... 317 3e-85
ref|NP_113453.1| photosystem II stability/assembly factor HCF136... 250 3e-65
ref|NP_682485.1| photosystem II stability/assembly factor [Therm... 218 1e-55
ref|NP_043177.1| unknown [Cyanophora paradoxa] gi|1351794|sp|P48... 213 3e-54
ref|ZP_00110640.1| COG4447: Uncharacterized protein related to p... 205 1e-51
>ref|NP_197703.1| photosystem II stability/assembly factor HCF136 [Arabidopsis
thaliana] gi|6016183|sp|O82660|H136_ARATH Photosystem II
stability/assembly factor HCF136, chloroplast precursor
gi|11358656|pir||T51828 probable photosystem II
stability protein HCF136 [imported] - Arabidopsis
thaliana gi|3559807|emb|CAA75723.1| HCF136 protein
[Arabidopsis thaliana] gi|9759370|dbj|BAB09829.1|
photosystem II stability/assembly factor HCF136
[Arabidopsis thaliana] gi|15010780|gb|AAK74049.1|
AT5g23120/MYJ24_11 [Arabidopsis thaliana]
gi|28416529|gb|AAO42795.1| At5g23120/MYJ24_11
[Arabidopsis thaliana]
Length = 403
Score = 317 bits (811), Expect = 3e-85
Identities = 170/276 (61%), Positives = 201/276 (72%), Gaps = 4/276 (1%)
Frame = +3
Query: 174 LATAAAVSMPIS-FPAPAQANQLLSTEWELVNLPVSQGTVLLDIGFTDDK--HGFLLGTR 344
L +AAVS+ +S PA+A++ LS EWE V LP+ G VLLDI F D+ GFLLGTR
Sbjct: 59 LYQSAAVSLSLSSIVGPARADEQLS-EWERVFLPIDPGVVLLDIAFVPDEPSRGFLLGTR 117
Query: 345 QTLLETNDGGKTWAPRSIDAARDEGFNYRFNSISFAGKEGWIVGKPAILLHTSDGGANWE 524
QTLLET DGG TW PRSI +A +E FNYRFNSISF GKEGWI+GKPAILL+T+D G NW+
Sbjct: 118 QTLLETKDGGSTWNPRSIPSAEEEDFNYRFNSISFKGKEGWIIGKPAILLYTADAGENWD 177
Query: 525 RIPLSSKLPGAPVRITGLAGKPGQAEMITDQGAIYVTDNTAYTWTAAVQETVDATLNRTV 704
RIPLSS+LPG V I K AEM+TD+GAIYVT N Y W AA+QETV ATLNRTV
Sbjct: 178 RIPLSSQLPGDMVFIKATEDK--SAEMVTDEGAIYVTSNRGYNWKAAIQETVSATLNRTV 235
Query: 705 SSGISGASYYEGSFSNLARSSSGDYVAVSSRGNFYMTWSPGQTYWMPHNPPRAPA-RLQN 881
SSGISGASYY G+FS + RS G YVAVSSRGNF++TW PGQ YW PHN RA A R+QN
Sbjct: 236 SSGISGASYYTGTFSAVNRSPDGRYVAVSSRGNFFLTWEPGQPYWQPHN--RAVARRIQN 293
Query: 882 MGFTPAGEVWVTTRGGDVLVSXRTPALGPARKFKDV 989
MG+ G +W+ RGG + +S T G +F++V
Sbjct: 294 MGWRADGGLWLLVRGGGLYLSKGT---GITEEFEEV 326
Score = 38.1 bits (87), Expect = 0.24
Identities = 25/82 (30%), Positives = 43/82 (51%), Gaps = 2/82 (2%)
Frame = +3
Query: 255 ELVNLPV-SQGTVLLDIGFTDDKHGFLLGTRQTLLETNDGGKTWAPRSIDAARDEGFNYR 431
E +PV S+G +LD+G+ ++ + G LL T +GGK+W + D A D
Sbjct: 322 EFEEVPVQSRGFGILDVGYRSEEEAWAAGGSGILLRTRNGGKSW---NRDKAAD-NIAAN 377
Query: 432 FNSISFA-GKEGWIVGKPAILL 494
++ F K+G+++G +LL
Sbjct: 378 LYAVKFVDDKKGFVLGNDGVLL 399
>ref|NP_113453.1| photosystem II stability/assembly factor HCF136 [Guillardia theta]
gi|25396851|pir||C90108 photosystem II
stability/assembly factor HCF136 [imported] - Guillardia
theta nucleomorph gi|12580704|emb|CAC27022.1|
photosystem II stability/assembly factor HCF136
[Guillardia theta]
Length = 395
Score = 250 bits (638), Expect = 3e-65
Identities = 127/253 (50%), Positives = 170/253 (66%), Gaps = 6/253 (2%)
Frame = +3
Query: 210 FPAPAQANQLLSTEWELVNLPVSQGTVLLDIGFTDD--KHGFLLGTRQTLLETNDGGKTW 383
+P AN +T W V+LPV +VL DI FTD KHG+L+G++ T LET+DGG TW
Sbjct: 60 YPKFTHAN---TTSWTKVDLPVD--SVLFDIEFTDPECKHGWLVGSKGTFLETDDGGNTW 114
Query: 384 APRSI-DAARDEGFNYRFNSISFAGKEGWIVGKPAILLHTSDGGANWERIPLSSKLPGAP 560
PR+ + DE YRF +ISF G+EGW++GKPAI+L+T DGG W R+P+S KLPG P
Sbjct: 115 VPRTFANLDPDEELTYRFENISFEGQEGWVIGKPAIILYTKDGGKTWFRVPVSPKLPGEP 174
Query: 561 VRITGLAGKPGQAEMITDQGAIYVTDNTAYTWTAAVQETVDATLNRTVSSGISGASYYEG 740
I L + AE+ T GAIYVT+N W A V+ET+D+TLNRT+SSG+SGASY+ G
Sbjct: 175 CLIKALGSE--SAELTTTSGAIYVTNNAGRNWKAQVKETIDSTLNRTISSGVSGASYFTG 232
Query: 741 SFSNLARSSSGDYVAVSSRGNFYMTWSPGQTYWMPHNPPRAPARLQNMGFTPAGE---VW 911
+ N+ R+S G Y+A+SSRGNFY+TW PGQ +W+P R+Q+MGF +W
Sbjct: 233 NVINVIRNSEGKYLAISSRGNFYLTWEPGQDFWIP-RARETSRRIQSMGFIQNDNQKGIW 291
Query: 912 VTTRGGDVLVSXR 950
++TRGG + VS +
Sbjct: 292 MSTRGGGLSVSTK 304
>ref|NP_682485.1| photosystem II stability/assembly factor [Thermosynechococcus
elongatus BP-1] gi|22295420|dbj|BAC09247.1| photosystem
II stability/assembly factor [Thermosynechococcus
elongatus BP-1]
Length = 347
Score = 218 bits (555), Expect = 1e-55
Identities = 117/248 (47%), Positives = 146/248 (58%), Gaps = 1/248 (0%)
Frame = +3
Query: 204 ISFPAPAQANQLLS-TEWELVNLPVSQGTVLLDIGFTDDKHGFLLGTRQTLLETNDGGKT 380
+S PA A L WE + LP + +LD+ F D HG+L+G TL+ET DGG+T
Sbjct: 30 VSLSTPALAIPALDYNPWEAIQLPTT--ATILDMSFIDRHHGWLVGVNATLMETRDGGQT 87
Query: 381 WAPRSIDAARDEGFNYRFNSISFAGKEGWIVGKPAILLHTSDGGANWERIPLSSKLPGAP 560
W PR++ + YRFNS+SF G EGWIVG+P I+LHT+DGG +W +IPL KLPG+P
Sbjct: 88 WEPRTLVLDHSD---YRFNSVSFQGNEGWIVGEPPIMLHTTDGGQSWSQIPLDPKLPGSP 144
Query: 561 VRITGLAGKPGQAEMITDQGAIYVTDNTAYTWTAAVQETVDATLNRTVSSGISGASYYEG 740
I L G AEMIT+ GAIY T ++ W A VQE + G
Sbjct: 145 RLIKALGN--GSAEMITNVGAIYRTKDSGKNWQALVQEAI-------------------G 183
Query: 741 SFSNLARSSSGDYVAVSSRGNFYMTWSPGQTYWMPHNPPRAPARLQNMGFTPAGEVWVTT 920
NL RS SG+YVAVSSRG+FY TW PGQT W PHN + RL NMGFTP G +W+
Sbjct: 184 VMRNLNRSPSGEYVAVSSRGSFYSTWEPGQTAWEPHNRTTS-RRLHNMGFTPDGRLWMIV 242
Query: 921 RGGDVLVS 944
GG + S
Sbjct: 243 NGGKIAFS 250
Score = 35.4 bits (80), Expect = 1.5
Identities = 44/162 (27%), Positives = 65/162 (39%), Gaps = 9/162 (5%)
Frame = +3
Query: 189 AVSMPISFPAPAQANQLLSTEWELVNLPVSQGTVLLDIGFTDD-KHGFLLGTRQTLLETN 365
AVS SF + + Q T WE N S+ L ++GFT D + ++ +
Sbjct: 198 AVSSRGSFYSTWEPGQ---TAWEPHNRTTSRR--LHNMGFTPDGRLWMIVNGGKIAFSDP 252
Query: 366 DGGKTWAPRSIDAARDEGFNYRFNSISFAG------KEGWIVGKPAILLHTSDGGANWER 527
D + W R NS+ F E W+ G LL + DGG W++
Sbjct: 253 DNSENWGELLSPLRR--------NSVGFLDLAYRTPNEVWLAGGAGALLCSQDGGQTWQQ 304
Query: 528 IPLSSKLPGAPVRITGLAGKPGQAEMITDQGAI--YVTDNTA 647
K+P +I L P Q ++ +G + YVTD TA
Sbjct: 305 DVDVKKVPSNFYKI--LFFSPDQGFILGQKGILLRYVTDLTA 344
>ref|NP_043177.1| unknown [Cyanophora paradoxa] gi|1351794|sp|P48325|YC48_CYAPA
Hypothetical 37.3 kDa protein ycf48 (ORF333)
gi|7484243|pir||T06865 hypothetical protein 333 -
Cyanophora paradoxa cyanelle gi|1016121|gb|AAA81208.1|
putative protein of 333 amino acids
Length = 333
Score = 213 bits (543), Expect = 3e-54
Identities = 114/236 (48%), Positives = 149/236 (62%), Gaps = 2/236 (0%)
Frame = +3
Query: 243 STEWELVNLPVSQGTVLLDIGFTDDK--HGFLLGTRQTLLETNDGGKTWAPRSIDAARDE 416
S +WE +P++ +LLDIGF D+ G+LLGTR TL ET D GKTW RS++ D+
Sbjct: 30 SYKWE--QIPLNTDEILLDIGFVPDQPQRGWLLGTRSTLFETTDKGKTWELRSLNLEDDK 87
Query: 417 GFNYRFNSISFAGKEGWIVGKPAILLHTSDGGANWERIPLSSKLPGAPVRITGLAGKPGQ 596
YR NSISF+GKEGW+ GKPAILLHT+DGG++W RIPLS++LPG P IT L G+
Sbjct: 88 ---YRLNSISFSGKEGWVTGKPAILLHTTDGGSSWSRIPLSNQLPGDPALITALG--TGK 142
Query: 597 AEMITDQGAIYVTDNTAYTWTAAVQETVDATLNRTVSSGISGASYYEGSFSNLARSSSGD 776
AE+ TD GAIY T+N+ TW A +QE + G +ARS +G
Sbjct: 143 AELATDIGAIYRTENSGQTWKAQIQEPL-------------------GVIRTVARSENGS 183
Query: 777 YVAVSSRGNFYMTWSPGQTYWMPHNPPRAPARLQNMGFTPAGEVWVTTRGGDVLVS 944
YVAVS++GNFY TW G W+ H P ++ R+Q+MGFT +W+ TRGG + S
Sbjct: 184 YVAVSAKGNFYSTWKEGDDKWISH-PRQSSRRIQSMGFTNNNRLWMLTRGGQLWFS 238
>ref|ZP_00110640.1| COG4447: Uncharacterized protein related to plant photosystem II
stability/assembly factor [Nostoc punctiforme]
Length = 339
Score = 205 bits (521), Expect = 1e-51
Identities = 107/235 (45%), Positives = 145/235 (61%), Gaps = 1/235 (0%)
Frame = +3
Query: 252 WELVNLPVSQGTVLLDIGFTDD-KHGFLLGTRQTLLETNDGGKTWAPRSIDAARDEGFNY 428
W+++++P LLDI FT++ +HG+L+G+ TLLETNDGG +W P ++ + Y
Sbjct: 35 WKVISVPTDSN--LLDIAFTNNPEHGYLVGSNATLLETNDGGNSWKPITLQLDEQK---Y 89
Query: 429 RFNSISFAGKEGWIVGKPAILLHTSDGGANWERIPLSSKLPGAPVRITGLAGKPGQAEMI 608
RF+S+SFAGKEGWI G+P +LLHT+D GA+W RIPLS KLPG+P+ + LA AEM
Sbjct: 90 RFDSVSFAGKEGWIAGEPGLLLHTTDEGASWSRIPLSEKLPGSPIAVKALA--DNTAEMA 147
Query: 609 TDQGAIYVTDNTAYTWTAAVQETVDATLNRTVSSGISGASYYEGSFSNLARSSSGDYVAV 788
T+ GAIY T + W A V+ V G NL RS+ G YVAV
Sbjct: 148 TNVGAIYKTTDGGKNWKAQVESAV-------------------GVVRNLERSADGKYVAV 188
Query: 789 SSRGNFYMTWSPGQTYWMPHNPPRAPARLQNMGFTPAGEVWVTTRGGDVLVSXRT 953
S++G+FY TW PGQ W+PHN + R++NMGF G++W+ RGG V S T
Sbjct: 189 SAKGSFYSTWEPGQDAWVPHN-RNSSRRVENMGFGDNGQLWLLARGGQVQFSDPT 242
Score = 33.1 bits (74), Expect = 7.6
Identities = 23/83 (27%), Positives = 38/83 (45%), Gaps = 1/83 (1%)
Frame = +3
Query: 249 EWELVNLP-VSQGTVLLDIGFTDDKHGFLLGTRQTLLETNDGGKTWAPRSIDAARDEGFN 425
EW+ P +S LLD+ + + ++ G LL + DGGKTW D +E
Sbjct: 246 EWQEAKYPELSTSWGLLDLAYRTPEEIWVGGGSGNLLRSADGGKTWEK---DRDIEEVAA 302
Query: 426 YRFNSISFAGKEGWIVGKPAILL 494
+ + + G+I+G +LL
Sbjct: 303 NLYKIVFLEPERGFIIGDRGVLL 325