Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001085A_C01 KCC001085A_c01
(882 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
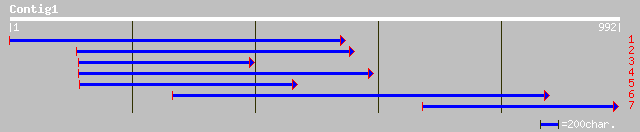
gb|AAM62459.1| unknown [Arabidopsis thaliana] 86 6e-16
ref|NP_567785.1| rhodanese-like domain protein [Arabidopsis thal... 86 6e-16
pir||T05875 hypothetical protein T29A15.190 - Arabidopsis thalia... 86 8e-16
dbj|BAC87331.1| unnamed protein product [Homo sapiens] 51 2e-05
ref|ZP_00007171.1| COG1663: Tetraacyldisaccharide-1-P 4'-kinase ... 49 8e-05
>gb|AAM62459.1| unknown [Arabidopsis thaliana]
Length = 224
Score = 86.3 bits (212), Expect = 6e-16
Identities = 59/192 (30%), Positives = 97/192 (49%), Gaps = 6/192 (3%)
Frame = +1
Query: 55 RPAHRVGRRSTVLVSAAAATTEMPR----WPVVFAKLTAAKVQSVSPEEAARRVESGEWL 222
R A V T+ + + P W L +V+SV +EA R + ++
Sbjct: 33 RSAAMVSGHKTLKIQCTSTKPAKPAAEVDWRQKRELLLEKRVRSVDVKEAQRLQKENNFV 92
Query: 223 LVDVRLAEQHQTGAPEGAVNVPIYETITMEGA-DFRKLLKAVMYKSNGV-NPVDPNPKFN 396
++DVR +++ G P GA+NV +Y I A D + L + G+ + + NP+F
Sbjct: 93 ILDVRPEAEYKAGHPPGAINVEMYRLIREWTAWDIARRLGFAFF---GIFSGTEENPEFI 149
Query: 397 EQIKAAVAKAGAKGVITMCEAGGTLKPSTNFPEGKPSRSLQAAYRVLTEGLAPSVAHLDR 576
+ ++A + K +I C + GT+KP+ N PEG+ SRSL AAY ++ G +V HL+
Sbjct: 150 QSVEAKLDKEAK--IIVACSSAGTMKPTQNLPEGQQSRSLIAAYLLVLNGY-KNVFHLEG 206
Query: 577 GVYGWYQADLPM 612
G+Y W + LP+
Sbjct: 207 GIYTWGKEGLPV 218
>ref|NP_567785.1| rhodanese-like domain protein [Arabidopsis thaliana]
gi|15215616|gb|AAK91353.1| AT4g27700/T29A15_190
[Arabidopsis thaliana] gi|20334878|gb|AAM16195.1|
AT4g27700/T29A15_190 [Arabidopsis thaliana]
Length = 224
Score = 86.3 bits (212), Expect = 6e-16
Identities = 59/192 (30%), Positives = 97/192 (49%), Gaps = 6/192 (3%)
Frame = +1
Query: 55 RPAHRVGRRSTVLVSAAAATTEMPR----WPVVFAKLTAAKVQSVSPEEAARRVESGEWL 222
R A V T+ + + P W L +V+SV +EA R + ++
Sbjct: 33 RSAAMVSGHKTLKIQCTSTKPAKPAAEVDWRQKRELLLEKRVRSVDVKEAQRLQKENNFV 92
Query: 223 LVDVRLAEQHQTGAPEGAVNVPIYETITMEGA-DFRKLLKAVMYKSNGV-NPVDPNPKFN 396
++DVR +++ G P GA+NV +Y I A D + L + G+ + + NP+F
Sbjct: 93 ILDVRPEAEYKAGHPPGAINVEMYRLIREWTAWDIARRLGFAFF---GIFSGTEENPEFI 149
Query: 397 EQIKAAVAKAGAKGVITMCEAGGTLKPSTNFPEGKPSRSLQAAYRVLTEGLAPSVAHLDR 576
+ ++A + K +I C + GT+KP+ N PEG+ SRSL AAY ++ G +V HL+
Sbjct: 150 QSVEAKLDKEAK--IIVACSSAGTMKPTQNLPEGQQSRSLIAAYLLVLNGY-KNVFHLEG 206
Query: 577 GVYGWYQADLPM 612
G+Y W + LP+
Sbjct: 207 GIYTWGKEGLPV 218
>pir||T05875 hypothetical protein T29A15.190 - Arabidopsis thaliana
gi|4469021|emb|CAB38282.1| hypothetical protein
[Arabidopsis thaliana] gi|7269624|emb|CAB81420.1|
hypothetical protein [Arabidopsis thaliana]
Length = 237
Score = 85.9 bits (211), Expect = 8e-16
Identities = 53/156 (33%), Positives = 88/156 (55%), Gaps = 2/156 (1%)
Frame = +1
Query: 151 LTAAKVQSVSPEEAARRVESGEWLLVDVRLAEQHQTGAPEGAVNVPIYETITMEGA-DFR 327
L +V+SV +EA R + ++++DVR +++ G P GA+NV +Y I A D
Sbjct: 82 LLEKRVRSVDVKEAQRLQKENNFVILDVRPEAEYKAGHPPGAINVEMYRLIREWTAWDIA 141
Query: 328 KLLKAVMYKSNGV-NPVDPNPKFNEQIKAAVAKAGAKGVITMCEAGGTLKPSTNFPEGKP 504
+ L + G+ + + NP+F + ++A + K +I C + GT+KP+ N PEG+
Sbjct: 142 RRLGFAFF---GIFSGTEENPEFIQSVEAKLDKEAK--IIVACSSAGTMKPTQNLPEGQQ 196
Query: 505 SRSLQAAYRVLTEGLAPSVAHLDRGVYGWYQADLPM 612
SRSL AAY ++ G +V HL+ G+Y W + LP+
Sbjct: 197 SRSLIAAYLLVLNGY-KNVFHLEGGIYTWGKEGLPV 231
>dbj|BAC87331.1| unnamed protein product [Homo sapiens]
Length = 516
Score = 51.2 bits (121), Expect = 2e-05
Identities = 66/217 (30%), Positives = 81/217 (36%), Gaps = 3/217 (1%)
Frame = +3
Query: 27 ARFSCSRASSSRPSGGSP---FHGAGIRGRCHHGDAALAGGVRQADRC*GPERLA*GGRS 197
AR + RA + P GGS +HGAG G CH G GG + + G + LA G +
Sbjct: 143 ARVAAGRACRAPPGGGSRGARYHGAG-GGGCHGGAGVRIGGQHRLEAVLGQQVLAGGEQ- 200
Query: 198 ARRVRGVAAGGCSPGGAAPDRCAGGCCERAHLRDHYHGGG*LPQVAEGCHVQVQRRKPGG 377
GG GGA GG ERA + G ++VQR G
Sbjct: 201 --------VGGGGGGGAEARGAEGGVRERAEVG------------VVGAQLRVQRGLGGA 240
Query: 378 PQPQVQRANQGSSGQGRRQGCHHHVRGGRHPQALHQLSRGQAQPLAPGGLPRPDRGPGPV 557
+VQRA + SG LH R +A P PR R G +
Sbjct: 241 ---RVQRALEPPSG------------------VLHFQPRARAAVHVP---PRRRRAVGHL 276
Query: 558 CGPSGSGRVRLVPGGSAHERRVQARHRPHAHGRRRAH 668
G RLVP G R AR +P G RR H
Sbjct: 277 AGRQRRMEARLVP-GLVRVRHAHARVQP--VGCRRVH 310
>ref|ZP_00007171.1| COG1663: Tetraacyldisaccharide-1-P 4'-kinase [Rhodobacter
sphaeroides]
Length = 590
Score = 49.3 bits (116), Expect = 8e-05
Identities = 74/267 (27%), Positives = 89/267 (32%), Gaps = 29/267 (10%)
Frame = +3
Query: 21 KNARFSCSRASSSRPSGGSPFHGAGIRGRCHHGDAALAGGVRQ-----ADRC*GPERLA* 185
+ R R +R +GG P G R H G A G + A R GP
Sbjct: 22 RRPRLPQGRKLLARHAGGGPARGGQPRAALHRGRARRVGAASRPPAGVARR--GPSHRGR 79
Query: 186 GGRSARRVRGVAAGGCSPGGAAPDRCAGGCCERAHLRDHYHGGG*L---PQVAEGC---H 347
G R R RG A A DR A G R R G G P+ G H
Sbjct: 80 GRRHRRASRGAQARAPD----AADRRAAGARPRRRFRRALRGRGLEGRPPRRRRGAGRRH 135
Query: 348 VQVQRRKPGGPQPQVQRANQGSSGQGRRQGCHHHVRGGRHPQALHQLSRGQAQPLAP--- 518
++ R+ GG +P V + G+ RRQ RG R A L+R P P
Sbjct: 136 RGLRGRRHGGARPLV---SAGAGDMARRQP----QRGLRAGPARGGLARFGPDPRPPFRP 188
Query: 519 ---------GGLPRPDRGPGPVCGPSGSGRVRLVPGGSAHERRV------QARHRPHAHG 653
GG P RG GP G R GG+A R+ HRP
Sbjct: 189 AWRDAGPALGGARLPSRGLGPRPRRGAGGSARPGSGGAAGPCRLGRGLGRNRGHRPDGGA 248
Query: 654 RRRAHPAAVSTSERGYEMRPEDKPVEA 734
RR A + P D P A
Sbjct: 249 RRTPRRAGGLMRPPAFWFTPPDSPALA 275