Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001068A_C01 KCC001068A_c01
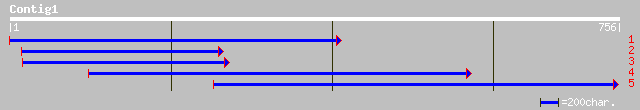
(756 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC28746.1| putative protein [Neurospora crassa] 43 0.006
ref|XP_323607.1| hypothetical protein [Neurospora crassa] gi|289... 43 0.006
gb|AAF14115.1| SPT6 protein [Drosophila melanogaster] 41 0.023
ref|NP_651962.2| CG12225-PA [Drosophila melanogaster] gi|7290693... 41 0.023
gb|AAF14114.1|AF104400_1 SPT6 protein [Drosophila melanogaster] 41 0.023
>emb|CAC28746.1| putative protein [Neurospora crassa]
Length = 600
Score = 42.7 bits (99), Expect = 0.006
Identities = 30/95 (31%), Positives = 44/95 (45%), Gaps = 4/95 (4%)
Frame = +2
Query: 467 ILEMLCKVAGAVLAWSLHRSSTQGEVPYQPVSSSAGAHHQGGAGGGSGAAGGYGQMPSAD 646
+L L +A ++ W +S++ G+ SS G+ G GGSG +GG G S
Sbjct: 311 LLFCLVLIAACIVDWFSTKSNSSGDRGRSSRSSGRGSRTSGSGSGGSGGSGGSGGGGSKR 370
Query: 647 PFAGYHRHSRTRCTRRC---PPR-TNLSPLASPHP 739
RHSR+ + + PPR +N P A P P
Sbjct: 371 SRKESRRHSRSPGSSKSSPGPPRPSNTFPPAPPPP 405
>ref|XP_323607.1| hypothetical protein [Neurospora crassa] gi|28922800|gb|EAA32022.1|
hypothetical protein [Neurospora crassa]
Length = 605
Score = 42.7 bits (99), Expect = 0.006
Identities = 30/95 (31%), Positives = 44/95 (45%), Gaps = 4/95 (4%)
Frame = +2
Query: 467 ILEMLCKVAGAVLAWSLHRSSTQGEVPYQPVSSSAGAHHQGGAGGGSGAAGGYGQMPSAD 646
+L L +A ++ W +S++ G+ SS G+ G GGSG +GG G S
Sbjct: 316 LLFCLVLIAACIVDWFSTKSNSSGDRGRSSRSSGRGSRTSGSGSGGSGGSGGSGGGGSKR 375
Query: 647 PFAGYHRHSRTRCTRRC---PPR-TNLSPLASPHP 739
RHSR+ + + PPR +N P A P P
Sbjct: 376 SRKESRRHSRSPGSSKSSPGPPRPSNTFPPAPPPP 410
>gb|AAF14115.1| SPT6 protein [Drosophila melanogaster]
Length = 1827
Score = 40.8 bits (94), Expect = 0.023
Identities = 25/70 (35%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +2
Query: 518 HRSSTQGEVPYQPVSSSAGAHHQGGAGGGSGAAGGYGQMPSAD--PFAGYHRHSRTRCTR 691
H S Q P+ +S + G G G G G GGYG P D G++R ++ +
Sbjct: 1746 HHSQQQ---PHMGMSMNMGITMSLGRGTGGGGGGGYGSTPVNDYSTGGGHNRGMSSKASV 1802
Query: 692 RCPPRTNLSP 721
R PRTN SP
Sbjct: 1803 RSTPRTNASP 1812
>ref|NP_651962.2| CG12225-PA [Drosophila melanogaster] gi|7290693|gb|AAF46140.1|
CG12225-PA [Drosophila melanogaster]
Length = 1831
Score = 40.8 bits (94), Expect = 0.023
Identities = 25/70 (35%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +2
Query: 518 HRSSTQGEVPYQPVSSSAGAHHQGGAGGGSGAAGGYGQMPSAD--PFAGYHRHSRTRCTR 691
H S Q P+ +S + G G G G G GGYG P D G++R ++ +
Sbjct: 1750 HHSQQQ---PHMGMSMNMGITMSLGRGTGGGGGGGYGSTPVNDYSTGGGHNRGMSSKASV 1806
Query: 692 RCPPRTNLSP 721
R PRTN SP
Sbjct: 1807 RSTPRTNASP 1816
>gb|AAF14114.1|AF104400_1 SPT6 protein [Drosophila melanogaster]
Length = 1831
Score = 40.8 bits (94), Expect = 0.023
Identities = 25/70 (35%), Positives = 33/70 (46%), Gaps = 2/70 (2%)
Frame = +2
Query: 518 HRSSTQGEVPYQPVSSSAGAHHQGGAGGGSGAAGGYGQMPSAD--PFAGYHRHSRTRCTR 691
H S Q P+ +S + G G G G G GGYG P D G++R ++ +
Sbjct: 1750 HHSQQQ---PHMGMSMNMGITMSLGRGTGGGGGGGYGSTPVNDYSTGGGHNRGMSSKASV 1806
Query: 692 RCPPRTNLSP 721
R PRTN SP
Sbjct: 1807 RSTPRTNASP 1816