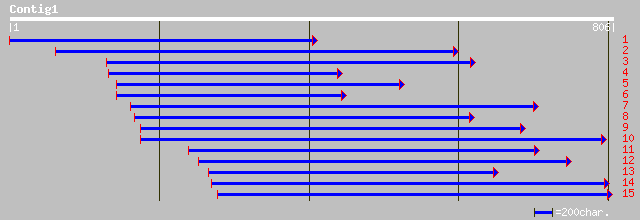
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC001049A_C01 KCC001049A_c01
(690 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|XP_301558.1| similar to Protein kinase PKX1 [Homo sapiens] 37 0.37
ref|XP_310616.1| ENSANGP00000020759 [Anopheles gambiae] gi|21294... 36 0.63
ref|XP_298502.1| hypothetical protein XP_298502 [Homo sapiens] 35 0.82
ref|NP_730262.2| CG13731-PA [Drosophila melanogaster] gi|2838049... 35 1.1
ref|ZP_00089282.1| COG0655: Multimeric flavodoxin WrbA [Azotobac... 35 1.4
>ref|XP_301558.1| similar to Protein kinase PKX1 [Homo sapiens]
Length = 1593
Score = 36.6 bits (83), Expect = 0.37
Identities = 23/64 (35%), Positives = 26/64 (39%), Gaps = 1/64 (1%)
Frame = +1
Query: 415 CCWWHSVCRYSSQLWLGHAQSGI*RPNTAPAGGLVTQPVTSHPPITHWSPTG-RPPVTHR 591
C W S R SS H RP + +P SHPP+ H P G PPV H
Sbjct: 1296 CVWKESSARSSSASGRNHPPVPHLRPEGSHPPVPHLRPEGSHPPVPHLRPEGSHPPVPHL 1355
Query: 592 PPTG 603
P G
Sbjct: 1356 RPEG 1359
Score = 33.5 bits (75), Expect = 3.1
Identities = 18/41 (43%), Positives = 21/41 (50%), Gaps = 1/41 (2%)
Frame = +1
Query: 487 RPNTAPAGGLVTQPVTSHPPITHWSPT-GRPPVTHRPPTGG 606
RP++A G SHPP+ H P G PPV H P GG
Sbjct: 710 RPSSASEG--------SHPPVPHLRPEGGHPPVPHLRPEGG 742
Score = 32.0 bits (71), Expect = 9.1
Identities = 24/69 (34%), Positives = 30/69 (42%), Gaps = 7/69 (10%)
Frame = +1
Query: 418 CWWHSVCRYSSQLWLGHAQSGI*RPNTA------PAGGLVTQPVTSHPPITHWSPTG-RP 576
C W V R S L + + RP++A P L + SHPP+ H P G P
Sbjct: 797 CVWKEVIRPSLICVLKESST---RPSSASGRNHPPLPHLRPEGEGSHPPVPHLRPEGSHP 853
Query: 577 PVTHRPPTG 603
PV H P G
Sbjct: 854 PVPHLRPEG 862
>ref|XP_310616.1| ENSANGP00000020759 [Anopheles gambiae] gi|21294177|gb|EAA06322.1|
ENSANGP00000020759 [Anopheles gambiae str. PEST]
Length = 683
Score = 35.8 bits (81), Expect = 0.63
Identities = 28/68 (41%), Positives = 32/68 (46%), Gaps = 4/68 (5%)
Frame = +2
Query: 209 PTTTCRAPSTRTA*LKGSPTTLYAACLPSLHHG----VLVRPRSWEVGLADSHGEGGS*G 376
PT R TR LKG ++ AA P G VL R W+VGL HG G G
Sbjct: 491 PTAFQRECFTRV--LKGF-LSVAAAVFPVRASGTTFDVLARKALWDVGLDYGHGTGHGIG 547
Query: 377 AFHGAHDY 400
AF G H+Y
Sbjct: 548 AFLGVHEY 555
>ref|XP_298502.1| hypothetical protein XP_298502 [Homo sapiens]
Length = 225
Score = 35.4 bits (80), Expect = 0.82
Identities = 21/61 (34%), Positives = 23/61 (37%), Gaps = 13/61 (21%)
Frame = -1
Query: 432 RVPPAAAAVLP*S*APWKAPQDPPSPWLSARPTSHE-------------RGRTRTPWWRL 292
R PP A P PW P PPSP S PT + RG+ PW R
Sbjct: 47 RSPPHAGTARPRPRGPWPCPAPPPSPRRSPGPTRLQSALRLRGRIRRKGRGQVALPWLRA 106
Query: 291 G 289
G
Sbjct: 107 G 107
>ref|NP_730262.2| CG13731-PA [Drosophila melanogaster] gi|28380492|gb|AAF49349.4|
CG13731-PA [Drosophila melanogaster]
Length = 926
Score = 35.0 bits (79), Expect = 1.1
Identities = 18/39 (46%), Positives = 22/39 (56%), Gaps = 2/39 (5%)
Frame = +1
Query: 490 PNTAPAGGLVTQPVTSHPPITHWSPTGR--PPVTHRPPT 600
P T P T+P T+ PP T+ PT + PPVT R PT
Sbjct: 320 PRTPPPTRPPTKPPTTRPPATYLPPTNKPPPPVTTRRPT 358
Score = 32.7 bits (73), Expect = 5.3
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Frame = +1
Query: 490 PNTAPAGGLVTQPVTSHPPITHWSPTGR--PPVTHRPP 597
P T P T+P T+ PP T+ PT + PPVT R P
Sbjct: 277 PRTPPPTRPPTRPPTTRPPATYLPPTNKPLPPVTTRLP 314
Score = 32.7 bits (73), Expect = 5.3
Identities = 17/38 (44%), Positives = 21/38 (54%), Gaps = 2/38 (5%)
Frame = +1
Query: 490 PNTAPAGGLVTQPVTSHPPITHWSPTGR--PPVTHRPP 597
P T P T+P T+ PP T+ PT + PPVT R P
Sbjct: 234 PRTPPPTRPPTRPPTTRPPATYLPPTNKPLPPVTTRLP 271
>ref|ZP_00089282.1| COG0655: Multimeric flavodoxin WrbA [Azotobacter vinelandii]
Length = 344
Score = 34.7 bits (78), Expect = 1.4
Identities = 22/49 (44%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Frame = +2
Query: 257 GSPTTLYAACLPSLHHGVLV--RPRSWEVGLADSHGEGGS*GAFHGAHD 397
G TTL + LP LHHG+L+ P S E L ++ G G GA H A D
Sbjct: 265 GQETTLLSMLLPLLHHGMLICGLPYS-EAALLETRGGGTPYGASHHAGD 312