Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
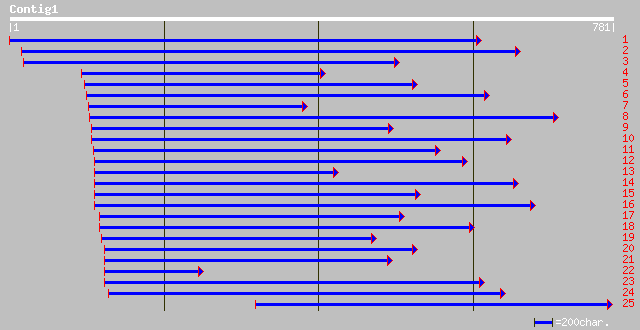
Query= KCC001043A_C01 KCC001043A_c01
(660 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
ref|ZP_00071811.1| COG0211: Ribosomal protein L27 [Trichodesmium... 64 6e-10
ref|NP_848958.1| ribosomal protein L27 [Cyanidioschyzon merolae]... 62 1e-09
ref|NP_484190.1| 50S ribosomal protein L27 [Nostoc sp. PCC 7120]... 63 2e-09
ref|NP_198911.1| 50S ribosomal protein L27, chloroplast precurso... 56 2e-09
gb|AAM62675.1| 50S ribosomal protein L27 [Arabidopsis thaliana] 55 3e-09
>ref|ZP_00071811.1| COG0211: Ribosomal protein L27 [Trichodesmium erythraeum IMS101]
Length = 95
Score = 64.3 bits (155), Expect(2) = 6e-10
Identities = 36/84 (42%), Positives = 59/84 (69%), Gaps = 3/84 (3%)
Frame = +1
Query: 139 KNGRDSNA*APRCEGLRRPG--LLKAGGIIVRQCGSTWNAGDNCMLGRDFTVFSTVEGVV 312
+NGRDSN+ + G++R G ++KAG I+VRQ GS ++AG+N +G+D+T+F+ VEG+V
Sbjct: 11 RNGRDSNS---QRLGVKRYGGQVVKAGNILVRQRGSKFHAGNNVGVGKDYTLFALVEGMV 67
Query: 313 VFDKK-RVKPEIHVYPADHPKAVA 381
F++K + + ++ VYP VA
Sbjct: 68 TFERKGKSRKKVSVYPLAQEANVA 91
Score = 21.6 bits (44), Expect(2) = 6e-10
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +3
Query: 111 AHKKGSGST 137
AHKKG+GST
Sbjct: 2 AHKKGTGST 10
>ref|NP_848958.1| ribosomal protein L27 [Cyanidioschyzon merolae]
gi|30409171|dbj|BAC76120.1| 50S ribosomal protein L27
[Cyanidioschyzon merolae]
Length = 104
Score = 62.0 bits (149), Expect(2) = 1e-09
Identities = 33/79 (41%), Positives = 49/79 (61%), Gaps = 2/79 (2%)
Frame = +1
Query: 139 KNGRDSNA*APRCEGLR--RPGLLKAGGIIVRQCGSTWNAGDNCMLGRDFTVFSTVEGVV 312
KNGRDSN P+ G++ L+ AG I+VRQ GS ++ G +DF++F+ ++G V
Sbjct: 11 KNGRDSN---PKYLGVKVGHAKLVSAGSILVRQRGSKFDPGMYVARAKDFSLFALIDGYV 67
Query: 313 VFDKKRVKPEIHVYPADHP 369
F +R KP IH++P HP
Sbjct: 68 QFAYRRGKPTIHIHPISHP 86
Score = 23.1 bits (48), Expect(2) = 1e-09
Identities = 9/10 (90%), Positives = 10/10 (100%)
Frame = +3
Query: 111 AHKKGSGSTE 140
AHKKGSGST+
Sbjct: 2 AHKKGSGSTK 11
>ref|NP_484190.1| 50S ribosomal protein L27 [Nostoc sp. PCC 7120]
gi|20139493|sp|Q8Z0F1|RL27_ANASP 50S ribosomal protein
L27 gi|25295481|pir||AB1825 50S ribosomal protein L27
[imported] - Nostoc sp. (strain PCC 7120)
gi|17135124|dbj|BAB77670.1| 50S ribosomal protein L27
[Nostoc sp. PCC 7120]
Length = 92
Score = 62.8 bits (151), Expect(2) = 2e-09
Identities = 37/85 (43%), Positives = 60/85 (70%), Gaps = 3/85 (3%)
Frame = +1
Query: 139 KNGRDSNA*APRCEGLRRPG--LLKAGGIIVRQCGSTWNAGDNCMLGRDFTVFSTVEGVV 312
+NGRDSNA + G++R G + AG I+VRQ G+ ++AG+N +G+D T+F+ V+GVV
Sbjct: 11 RNGRDSNA---QRLGVKRFGGQAVIAGNILVRQRGTKFHAGNNVGIGKDDTLFALVDGVV 67
Query: 313 VFDKK-RVKPEIHVYPADHPKAVAA 384
F++K + + ++ VYPA +AVA+
Sbjct: 68 TFERKGKSRKKVSVYPAAAAEAVAS 92
Score = 21.6 bits (44), Expect(2) = 2e-09
Identities = 8/9 (88%), Positives = 9/9 (99%)
Frame = +3
Query: 111 AHKKGSGST 137
AHKKG+GST
Sbjct: 2 AHKKGTGST 10
>ref|NP_198911.1| 50S ribosomal protein L27, chloroplast precursor (CL27)
[Arabidopsis thaliana] gi|20139726|sp|Q9FLN4|RK27_ARATH
50S ribosomal protein L27, chloroplast precursor
gi|9759141|dbj|BAB09697.1| 50S ribosomal protein L27
[Arabidopsis thaliana] gi|15028249|gb|AAK76713.1|
putative 50S ribosomal protein L27 [Arabidopsis
thaliana] gi|21436345|gb|AAM51342.1| putative 50S
ribosomal protein L27 [Arabidopsis thaliana]
Length = 198
Score = 56.2 bits (134), Expect(2) = 2e-09
Identities = 41/125 (32%), Positives = 69/125 (54%), Gaps = 7/125 (5%)
Frame = +1
Query: 139 KNGRDSNA*APRCEGLRRPG--LLKAGGIIVRQCGSTWNAGDNCMLGRDFTVFSTVEGVV 312
KNGRDS + G++ G + K G IIVRQ G+ ++AG N +G+D T+FS ++G+V
Sbjct: 68 KNGRDSPG---QRLGVKIYGDQVAKPGAIIVRQRGTKFHAGKNVGIGKDHTIFSLIDGLV 124
Query: 313 VFDK-KRVKPEIHVYP----ADHPKAVAALAKTHTKQPAEGQQSRAERRKATYTSRKATL 477
F+K + +I VYP ++P + A + + + E +++R E T + + L
Sbjct: 125 KFEKFGPDRKKISVYPREIVPENPNSYRARKRENFRLQREKKKARRENYSYTLPTPELVL 184
Query: 478 SVAQV 492
+ A V
Sbjct: 185 ASASV 189
Score = 27.3 bits (59), Expect(2) = 2e-09
Identities = 13/22 (59%), Positives = 17/22 (77%)
Frame = +3
Query: 75 APAPKRALVIQNAHKKGSGSTE 140
AP P L I++AHKKG+GST+
Sbjct: 50 APIP---LTIESAHKKGAGSTK 68
>gb|AAM62675.1| 50S ribosomal protein L27 [Arabidopsis thaliana]
Length = 196
Score = 55.5 bits (132), Expect(2) = 3e-09
Identities = 40/125 (32%), Positives = 69/125 (55%), Gaps = 7/125 (5%)
Frame = +1
Query: 139 KNGRDSNA*APRCEGLRRPG--LLKAGGIIVRQCGSTWNAGDNCMLGRDFTVFSTVEGVV 312
KNGRDS + G++ G + K G IIVRQ G+ ++AG N +G+D T+FS ++G+V
Sbjct: 66 KNGRDSPG---QRLGVKIYGDQVAKPGAIIVRQRGTKFHAGKNVGIGKDHTIFSLIDGLV 122
Query: 313 VFDK-KRVKPEIHVYP----ADHPKAVAALAKTHTKQPAEGQQSRAERRKATYTSRKATL 477
F+K + +I VYP ++P + A + + + E +++R E T + + L
Sbjct: 123 KFEKFGPDRKKISVYPREIVPENPNSYRARKRENFRLQREKKKARRENYTYTLPTPELVL 182
Query: 478 SVAQV 492
+ A +
Sbjct: 183 ASASI 187
Score = 27.7 bits (60), Expect(2) = 3e-09
Identities = 13/22 (59%), Positives = 17/22 (77%)
Frame = +3
Query: 75 APAPKRALVIQNAHKKGSGSTE 140
AP P L I++AHKKG+GST+
Sbjct: 48 APVP---LTIESAHKKGAGSTK 66