Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000974A_C01 KCC000974A_c01
(683 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
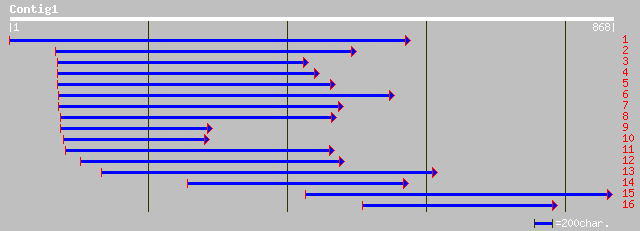
Sequences producing significant alignments: (bits) Value
gb|AAG02503.1|AF295371_1 chlamyopsin 2 [Chlamydomonas reinhardtii] 353 1e-96
pir||S60158 chlamyopsin - Chlamydomonas reinhardtii gi|1122270|e... 353 1e-96
pir||T10808 volvoxopsin - Volvox carteri gi|1914803|emb|CAA72088... 249 3e-65
ref|XP_311129.1| ENSANGP00000004749 [Anopheles gambiae] gi|30178... 49 5e-05
gb|AAQ82688.1| Epa4p [Candida glabrata] 48 2e-04
>gb|AAG02503.1|AF295371_1 chlamyopsin 2 [Chlamydomonas reinhardtii]
Length = 243
Score = 353 bits (906), Expect = 1e-96
Identities = 186/200 (93%), Positives = 188/200 (94%)
Frame = +3
Query: 6 MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK 185
MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK
Sbjct: 1 MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK 60
Query: 186 DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK 365
DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK
Sbjct: 61 DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK 120
Query: 366 AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLSCQVQTGLPCPSLIH 545
AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLS + PSLIH
Sbjct: 121 AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLSAKYSQAAG-PSLIH 179
Query: 546 EVEPSNEVLSLKSTCVCIKA 605
EVEPS +V SLKST V IKA
Sbjct: 180 EVEPSKKV-SLKST-VGIKA 197
>pir||S60158 chlamyopsin - Chlamydomonas reinhardtii gi|1122270|emb|CAA88832.1|
chlamyopsin [Chlamydomonas reinhardtii]
gi|4467383|emb|CAB37649.1| chlamyopsin [Chlamydomonas
reinhardtii] gi|1586088|prf||2203264A chlamyrhodopsin
Length = 235
Score = 353 bits (906), Expect = 1e-96
Identities = 186/200 (93%), Positives = 188/200 (94%)
Frame = +3
Query: 6 MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK 185
MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK
Sbjct: 1 MDFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSK 60
Query: 186 DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK 365
DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK
Sbjct: 61 DFSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARK 120
Query: 366 AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLSCQVQTGLPCPSLIH 545
AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLS + PSLIH
Sbjct: 121 AKLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLSAKYSQAAG-PSLIH 179
Query: 546 EVEPSNEVLSLKSTCVCIKA 605
EVEPS +V SLKST V IKA
Sbjct: 180 EVEPSKKV-SLKST-VGIKA 197
>pir||T10808 volvoxopsin - Volvox carteri gi|1914803|emb|CAA72088.1| volvoxopsin
[Volvox carteri] gi|1914805|emb|CAA93259.1| volvoxopsin
[Volvox carteri]
Length = 244
Score = 249 bits (636), Expect = 3e-65
Identities = 128/199 (64%), Positives = 154/199 (77%)
Frame = +3
Query: 9 DFKSISGEYDVSKKTLKTACKLKVADNISVKLKLANPGAKPGIEVEYKGFEATYDVKSKD 188
D K++ EYDV+KKTLK++CK KVAD++ VKLKL +P +EV+ K + TYDV KD
Sbjct: 3 DLKNLGAEYDVTKKTLKSSCKFKVADSVGVKLKLTSPANALNVEVDGKNWGITYDVVKKD 62
Query: 189 FSIEKKFKLRAGELKIKQKVPGLRTELLPSPEVQWKSHIVKGRKFSWEVEPSYCFQARKA 368
++ ++KL+ GE KIKQKVPGL+TELLPSPEVQWK H+VK +KF+WEVEPSYCFQAR A
Sbjct: 63 LELKGEWKLKGGEFKIKQKVPGLKTELLPSPEVQWKQHVVKNKKFAWEVEPSYCFQARLA 122
Query: 369 KLDQVIELNGGKEKLKLEFDSKLGVKGLVGTLTHKVGQSWAKQLSCQVQTGLPCPSLIHE 548
KL+Q +E NGGK+KLKLE DSK GVK VGTL+ KVG SWAKQLS + PSL HE
Sbjct: 123 KLEQTVEFNGGKQKLKLETDSKAGVKATVGTLSAKVGNSWAKQLSVKYSKAAG-PSLTHE 181
Query: 549 VEPSNEVLSLKSTCVCIKA 605
VEPSN+V SLKST V IKA
Sbjct: 182 VEPSNKV-SLKST-VGIKA 198
>ref|XP_311129.1| ENSANGP00000004749 [Anopheles gambiae] gi|30178092|gb|EAA06612.2|
ENSANGP00000004749 [Anopheles gambiae str. PEST]
Length = 633
Score = 49.3 bits (116), Expect = 5e-05
Identities = 43/131 (32%), Positives = 59/131 (44%), Gaps = 4/131 (3%)
Frame = -3
Query: 537 ARGTAGLSVLGSSAAWPRTGLPCASGCPPGPS--RPACC--RTPA*ASPCRR*AQSPGPA 370
A A + + A P T PCA+ CP PS RP+ C R+P A P R A P
Sbjct: 503 AAAAAAPAAVAVRPASPCTPSPCAAACPTSPSAARPSGCPGRSPPSAPPAARPAGPPAAC 562
Query: 369 WPCGPGSSSWAPPPTRTSCP*QCATSTGPRARAAARSSDRAPSA*SSARQPSA*TSSRC* 190
P S + PP+R +S GP A A+ S SA +AR P++ + R
Sbjct: 563 TARAPPGRSRSAPPSR--------SSAGP-AVASGCGSPPDCSASCAARPPASPAAPRSS 613
Query: 189 SPCSSRRRSPR 157
+P + R +PR
Sbjct: 614 APPGASRPAPR 624
>gb|AAQ82688.1| Epa4p [Candida glabrata]
Length = 1416
Score = 47.8 bits (112), Expect = 2e-04
Identities = 45/150 (30%), Positives = 76/150 (50%)
Frame = +1
Query: 112 PTLVPSQALRSSTRASRRPTT*RARTSASRRSSS*GLAS*RSSRRCPV*GPSCCPRPRSS 291
P PSQ + SS+ +S P++ + +S+S SSS +S SS S P P SS
Sbjct: 321 PLPTPSQDINSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSS 380
Query: 292 GSRTLSRAGSSRGRWSPATASRPARPSWTR*LSSTAARRSSSWSSTASWA*RAWWAP*RT 471
S + S + SS SP+ +S + S + SS+++ SSS SS++S + + +P +
Sbjct: 381 SSSSSSSSSSS----SPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSS 436
Query: 472 R*ASPGPSS*AAKYRQACRAPRSSTRWSPA 561
+S SS ++ + + SS+ SP+
Sbjct: 437 SSSSSSSSSSSSSSSSSSSSSSSSSSSSPS 466
Score = 44.7 bits (104), Expect = 0.001
Identities = 42/139 (30%), Positives = 69/139 (49%)
Frame = +1
Query: 10 TSRASAASMTSPRRR*RPPAS*RWPTTSPSSSSWPTLVPSQALRSSTRASRRPTT*RART 189
+S +S++S +S +S ++S SSS P+ S + SS+ +S P++ + +
Sbjct: 343 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSPSPSSSSSSS 402
Query: 190 SASRRSSS*GLAS*RSSRRCPV*GPSCCPRPRSSGSRTLSRAGSSRGRWSPATASRPARP 369
S+S SSS +S SS S P P SS S + S + SS S +++S +
Sbjct: 403 SSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSS 462
Query: 370 SWTR*LSSTAARRSSSWSS 426
S SS+++ SSS SS
Sbjct: 463 SSPSPSSSSSSSSSSSSSS 481
Score = 40.8 bits (94), Expect = 0.019
Identities = 41/139 (29%), Positives = 68/139 (48%)
Frame = +1
Query: 82 PTTSPSSSSWPTLVPSQALRSSTRASRRPTT*RARTSASRRSSS*GLAS*RSSRRCPV*G 261
P++S SSSS + S + SS+ +S ++ + + + SSS +S SS P
Sbjct: 339 PSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSPSPSSS 398
Query: 262 PSCCPRPRSSGSRTLSRAGSSRGRWSPATASRPARPSWTR*LSSTAARRSSSWSSTASWA 441
S SS S + S + SS S +++S P+ S + SS+++ SSS SS++S +
Sbjct: 399 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSS 458
Query: 442 *RAWWAP*RTR*ASPGPSS 498
+ +SP PSS
Sbjct: 459 --------SSSSSSPSPSS 469
Score = 38.1 bits (87), Expect = 0.12
Identities = 35/116 (30%), Positives = 57/116 (48%)
Frame = +1
Query: 88 TSPSSSSWPTLVPSQALRSSTRASRRPTT*RARTSASRRSSS*GLAS*RSSRRCPV*GPS 267
+S SSS P+ S + SS+ +S ++ + +S+S SSS + SS S
Sbjct: 331 SSSSSSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSS 390
Query: 268 CCPRPRSSGSRTLSRAGSSRGRWSPATASRPARPSWTR*LSSTAARRSSSWSSTAS 435
P P SS S + S + SS S +++S + S + S ++ SSS SS++S
Sbjct: 391 SSPSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSS 446
Score = 38.1 bits (87), Expect = 0.12
Identities = 37/118 (31%), Positives = 60/118 (50%)
Frame = +1
Query: 82 PTTSPSSSSWPTLVPSQALRSSTRASRRPTT*RARTSASRRSSS*GLAS*RSSRRCPV*G 261
P+ S SSSS + S + SS+ +S ++ + +S+ SSS +S SS P
Sbjct: 337 PSPSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSPSPS 396
Query: 262 PSCCPRPRSSGSRTLSRAGSSRGRWSPATASRPARPSWTR*LSSTAARRSSSWSSTAS 435
S SS S + S + SS S +++S + PS + SS+++ SSS SS++S
Sbjct: 397 SSSSSSSSSSSSSSSSSSSSSSSS-SSSSSSSSSSPSPSSSSSSSSSSSSSSSSSSSS 453
Score = 35.0 bits (79), Expect = 1.1
Identities = 38/121 (31%), Positives = 59/121 (48%)
Frame = +1
Query: 10 TSRASAASMTSPRRR*RPPAS*RWPTTSPSSSSWPTLVPSQALRSSTRASRRPTT*RART 189
+S +S++S +SP +S ++S SSSS + S + SS+ +S P+ + +
Sbjct: 382 SSSSSSSSSSSPSPSSSSSSS---SSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSS 438
Query: 190 SASRRSSS*GLAS*RSSRRCPV*GPSCCPRPRSSGSRTLSRAGSSRGRWSPATASRPARP 369
S+S SSS +S SS S P P SS S + S + SS S +S+P P
Sbjct: 439 SSSSSSSSSSSSSSSSSSSSS--SSSSSPSPSSSSSSSSSSSSSSP---SIQPSSKPVDP 493
Query: 370 S 372
S
Sbjct: 494 S 494
Score = 34.7 bits (78), Expect = 1.4
Identities = 40/139 (28%), Positives = 67/139 (47%)
Frame = +1
Query: 10 TSRASAASMTSPRRR*RPPAS*RWPTTSPSSSSWPTLVPSQALRSSTRASRRPTT*RART 189
+S +S++S +S +S ++S SSSS P+ S + SS+ +S P+ + +
Sbjct: 341 SSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSSPSPSSSSSSSSSSSSSSSPSPSSSSS 400
Query: 190 SASRRSSS*GLAS*RSSRRCPV*GPSCCPRPRSSGSRTLSRAGSSRGRWSPATASRPARP 369
S+S SSS +S SS SS S + S + SS S +++S +
Sbjct: 401 SSSSSSSSSSSSSSSSS---------------SSSSSSSSSSSSSPSPSSSSSSSSSSSS 445
Query: 370 SWTR*LSSTAARRSSSWSS 426
S + SS+++ SSS SS
Sbjct: 446 SSSSSSSSSSSSSSSSSSS 464