Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000903A_C01 KCC000903A_c01
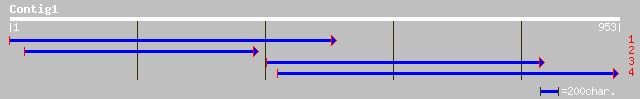
(953 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T08038 probable mitochondrial matrix protein precursor - Ch... 479 e-134
emb|CAE03469.1| OSJNBa0083N12.6 [Oryza sativa (japonica cultivar... 55 2e-06
ref|ZP_00050615.1| COG0523: Putative GTPases (G3E family) [Magne... 52 1e-05
gb|AAG31652.1| PRLI-interacting factor L [Arabidopsis thaliana] 50 4e-05
ref|NP_173025.1| PRLI-interacting factor L -related [Arabidopsis... 50 4e-05
>pir||T08038 probable mitochondrial matrix protein precursor - Chlamydomonas
reinhardtii gi|1877183|emb|CAA72325.1| putative
mitochondrial matrix protein [Chlamydomonas reinhardtii]
Length = 435
Score = 479 bits (1234), Expect = e-134
Identities = 225/248 (90%), Positives = 228/248 (91%)
Frame = +1
Query: 103 GGGWTRRR*RRGSARACGSRRRPAGRRSATLTRSCPSMSIRRRARRRGCVLRRQHKCTAS 282
G G + + G + PAGRRSATLTRSCPSMSIRRRARRRGCVLRRQHKCTAS
Sbjct: 188 GRGLDKEALKEGLSSCLWKPPPPAGRRSATLTRSCPSMSIRRRARRRGCVLRRQHKCTAS 247
Query: 283 MPRCPVRHAGAALRLGGCWGHAAPWEAHNGAFNFAMKALSLDCLALREQWNTDYEWGKCG 462
M PVRHAGAALRLGGCWGH APWEAHNGAFNFAMKALSLDCLALREQWNTDYEWGKCG
Sbjct: 248 MRAAPVRHAGAALRLGGCWGHPAPWEAHNGAFNFAMKALSLDCLALREQWNTDYEWGKCG 307
Query: 463 CARSRTQGQGAAFEAGLMVRAIHQVTVPNAGMRCICAPSSTCALQSDAGLHVYHALCLTI 642
CARSRTQGQGAAFEAGLMVRAIHQVTVPNAGMRCICAPSSTCALQSDAGLHVYHALCLTI
Sbjct: 308 CARSRTQGQGAAFEAGLMVRAIHQVTVPNAGMRCICAPSSTCALQSDAGLHVYHALCLTI 367
Query: 643 WTRSLVAARRRLSLLVCRDARCTNETGWGASDTWLLWLPGGLRADHSTCVASWHSYNSFH 822
WTRSLVAARRRLSLLVCRDARCTNETGWGASDTWLLWLPGGLRADHSTCVASWHSYNSFH
Sbjct: 368 WTRSLVAARRRLSLLVCRDARCTNETGWGASDTWLLWLPGGLRADHSTCVASWHSYNSFH 427
Query: 823 SCIRPWAC 846
SCIRPWAC
Sbjct: 428 SCIRPWAC 435
Score = 126 bits (317), Expect = 5e-28
Identities = 56/56 (100%), Positives = 56/56 (100%)
Frame = +3
Query: 3 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCLWKPPPP 170
KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCLWKPPPP
Sbjct: 155 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCLWKPPPP 210
>emb|CAE03469.1| OSJNBa0083N12.6 [Oryza sativa (japonica cultivar-group)]
Length = 804
Score = 55.1 bits (131), Expect = 2e-06
Identities = 25/50 (50%), Positives = 35/50 (70%)
Frame = +3
Query: 3 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCL 152
+FVF+G H + PA+ PW+PDE RF+ +VFIGR LD+ AL++ CL
Sbjct: 317 RFVFQGVHSMLEGCPAK-PWEPDEKRFNKLVFIGRNLDEAALRKAFKGCL 365
>ref|ZP_00050615.1| COG0523: Putative GTPases (G3E family) [Magnetospirillum
magnetotacticum]
Length = 343
Score = 52.4 bits (124), Expect = 1e-05
Identities = 26/49 (53%), Positives = 32/49 (65%)
Frame = +3
Query: 3 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSC 149
+FVF+G H I G + W DETR S VVFIGR LD EA++EG +C
Sbjct: 294 RFVFQGVH-MILDGDLQGAWGVDETRVSRVVFIGRNLDPEAIREGFYAC 341
>gb|AAG31652.1| PRLI-interacting factor L [Arabidopsis thaliana]
Length = 245
Score = 50.4 bits (119), Expect = 4e-05
Identities = 23/50 (46%), Positives = 35/50 (70%)
Frame = +3
Query: 3 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCL 152
+FVF+G HE I G ++ W+ DETR + +VFIG+ L++E L+ G +CL
Sbjct: 196 RFVFQGVHE-IFEGSPDRLWRKDETRTNKIVFIGKNLNREELEMGFRACL 244
>ref|NP_173025.1| PRLI-interacting factor L -related [Arabidopsis thaliana]
gi|25518551|pir||E86291 hypothetical protein F7H2.7
[imported] - Arabidopsis thaliana
gi|8927652|gb|AAF82143.1|AC034256_7 Contains similarity
to COBW-like protein from Homo sapiens gb|AF257330 and
contains a Viral (Superfamily 1) RNA helicase PF|01443
domain. EST gb|AI997977 comes from this genes.
[Arabidopsis thaliana]
gi|14194111|gb|AAK56250.1|AF367261_1 At1g15730/F7H2_7
[Arabidopsis thaliana] gi|20334730|gb|AAM16226.1|
At1g15730/F7H2_7 [Arabidopsis thaliana]
gi|23397243|gb|AAN31903.1| putative PRLI-interacting
factor L [Arabidopsis thaliana]
Length = 448
Score = 50.4 bits (119), Expect = 4e-05
Identities = 23/50 (46%), Positives = 35/50 (70%)
Frame = +3
Query: 3 KFVFKGAHEAICYGPAEQPWKPDETRFSHVVFIGRGLDKEALKEGLSSCL 152
+FVF+G HE I G ++ W+ DETR + +VFIG+ L++E L+ G +CL
Sbjct: 399 RFVFQGVHE-IFEGSPDRLWRKDETRTNKIVFIGKNLNREELEMGFRACL 447