Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000893A_C01 KCC000893A_c01
(893 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
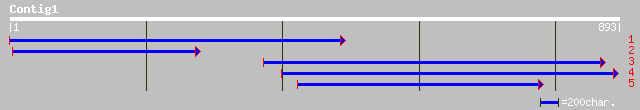
ref|ZP_00091451.1| COG3490: Uncharacterized protein conserved in... 38 0.26
ref|NP_294326.1| cytidine/deoxycytidylate deaminase/nudix/methyl... 33 4.9
emb|CAE02398.1| OSJNBa0024J22.2 [Oryza sativa (japonica cultivar... 33 4.9
ref|XP_352981.1| hypothetical protein XP_352980 [Homo sapiens] 33 6.4
>ref|ZP_00091451.1| COG3490: Uncharacterized protein conserved in bacteria [Azotobacter
vinelandii]
Length = 540
Score = 37.7 bits (86), Expect = 0.26
Identities = 50/166 (30%), Positives = 66/166 (39%), Gaps = 8/166 (4%)
Frame = -3
Query: 645 GNDRRDYS--SHQTREPKV---HGGSSRASKRASTTAGLLFRYLVWRAREPRNTPCCGRA 481
G+ RRD + H+ R P+ H RA+T AG + V A EPR P GRA
Sbjct: 24 GSHRRDPARRGHRPRCPEEEARHTARPAEQGRAATVAGRVLAQPV-HAFEPRRQPGDGRA 82
Query: 480 ALSSRRLKRRPRHAV*EATPGPG---P*SDVAMRL*APRLGLPSDDAHRTTERECFPDPS 310
++ R+ R P+ A GPG P R P+ RT R+ P
Sbjct: 83 PVARRQAGRHPQ----PARRGPGGTRPAPRPGFRRQPRAARRPAASVRRTARRQGRTRPV 138
Query: 309 EWPLHQSSLQHIVCPVLVTHPAAGGQLCPRVA*SAVQDMQCEKRRL 172
E PL Q + P + A GQ A +QC +RRL
Sbjct: 139 ERPLRQ------LRPPAPSARARAGQGARHPA-----RLQCPRRRL 173
>ref|NP_294326.1| cytidine/deoxycytidylate deaminase/nudix/methyltransferase domains
protein [Deinococcus radiodurans] gi|7471772|pir||C75499
cytidine/deoxycytidylate
deaminase/nudix/methyltransferase domains protein -
Deinococcus radiodurans (strain R1)
gi|6458301|gb|AAF10182.1|AE001918_7
cytidine/deoxycytidylate
deaminase/nudix/methyltransferase domains protein
[Deinococcus radiodurans]
Length = 548
Score = 33.5 bits (75), Expect = 4.9
Identities = 23/57 (40%), Positives = 28/57 (48%), Gaps = 1/57 (1%)
Frame = +2
Query: 101 GVIGGQAKSRRLQAHSNVT-LRSRPRRRFSHCMSWTAL*ATRGQSCPPAAGCVTNTG 268
GVIGG AH+ + L S P R C+SWT L T + CP AG V +G
Sbjct: 55 GVIGGHDL-----AHAEINALLSVPDLRRPECLSWTVL--TTVEPCPQCAGAVAMSG 104
>emb|CAE02398.1| OSJNBa0024J22.2 [Oryza sativa (japonica cultivar-group)]
gi|32492363|emb|CAE05121.1| OSJNBa0023J03.13 [Oryza
sativa (japonica cultivar-group)]
Length = 511
Score = 33.5 bits (75), Expect = 4.9
Identities = 19/66 (28%), Positives = 29/66 (43%)
Frame = -3
Query: 636 RRDYSSHQTREPKVHGGSSRASKRASTTAGLLFRYLVWRAREPRNTPCCGRAALSSRRLK 457
RR + H T P+ SS ++ S F+ +W+ R P P GR SRR +
Sbjct: 245 RRCRTRHSTASPRSTARSSTSASVPSARTARTFQESLWKWRPPPPPPSTGRPRSPSRRRR 304
Query: 456 RRPRHA 439
R++
Sbjct: 305 TTTRYS 310
>ref|XP_352981.1| hypothetical protein XP_352980 [Homo sapiens]
Length = 235
Score = 33.1 bits (74), Expect = 6.4
Identities = 28/81 (34%), Positives = 31/81 (37%), Gaps = 2/81 (2%)
Frame = -3
Query: 648 AGNDRRDYSSHQTREPKVHGGSSRASKRASTTAGLLFRYLVWRA--REPRNTPCCGRAAL 475
A RR + + EP G A AS T GL + L W R PR PC A
Sbjct: 109 AARSRRRWPPIRAAEPPARAGGVAAGS-ASATGGLGWAGLGWAGWGRPPRTHPCAAGPAW 167
Query: 474 SSRRLKRRPRHAV*EATPGPG 412
S RR ATPG G
Sbjct: 168 SPRR---------GSATPGSG 179
Database: nr
Posted date: Oct 14, 2003 12:26 AM
Number of letters in database: 498,525,298
Number of sequences in database: 1,537,769
Lambda K H
0.318 0.135 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 949,794,271
Number of Sequences: 1537769
Number of extensions: 22678649
Number of successful extensions: 70684
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 53259
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 70594
length of database: 498,525,298
effective HSP length: 123
effective length of database: 309,379,711
effective search space used: 53832069714
frameshift window, decay const: 50, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)