Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000773A_C01 KCC000773A_c01
(974 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
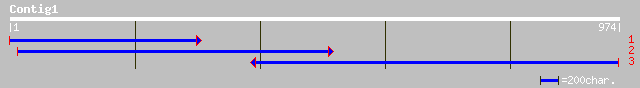
ref|XP_310365.1| ENSANGP00000018888 [Anopheles gambiae] gi|30178... 37 0.51
ref|XP_307017.1| ENSANGP00000016443 [Anopheles gambiae] gi|30179... 35 2.5
ref|ZP_00006349.1| COG1215: Glycosyltransferases, probably invol... 34 4.3
ref|XP_318131.1| ENSANGP00000017542 [Anopheles gambiae] gi|30175... 33 5.6
ref|XP_317932.1| ENSANGP00000011364 [Anopheles gambiae] gi|30175... 33 5.6
>ref|XP_310365.1| ENSANGP00000018888 [Anopheles gambiae] gi|30178154|gb|EAA06080.2|
ENSANGP00000018888 [Anopheles gambiae str. PEST]
Length = 399
Score = 37.0 bits (84), Expect = 0.51
Identities = 26/78 (33%), Positives = 35/78 (44%)
Frame = -2
Query: 931 QLLRLVRRLSVCRKCACHTVHSIIQYLKHYRSTVSCTPTKEGPCCLEQLLKPLQVPRHHV 752
Q L+ + RL +CRKC + HSI Q YR V+ + L Q++ P V + H
Sbjct: 194 QRLQTIERLKLCRKCLSN--HSIEQCKSRYRCYVNGCNKDDHHTLLHQVVVPQAVIQFHE 251
Query: 751 STSCAP*RLNSRVPCTAY 698
A L VP T Y
Sbjct: 252 KNKPAKSTLFKVVPVTLY 269
>ref|XP_307017.1| ENSANGP00000016443 [Anopheles gambiae] gi|30179409|gb|EAA02816.2|
ENSANGP00000016443 [Anopheles gambiae str. PEST]
Length = 427
Score = 34.7 bits (78), Expect = 2.5
Identities = 25/78 (32%), Positives = 35/78 (44%)
Frame = -2
Query: 931 QLLRLVRRLSVCRKCACHTVHSIIQYLKHYRSTVSCTPTKEGPCCLEQLLKPLQVPRHHV 752
Q L+ V RL +CRKC + HSI Q YR ++ + L Q+ P + + H
Sbjct: 152 QRLQTVERLKLCRKCLSN--HSIEQCKTRYRCYINGCHKDDHHTLLHQVDVPQAIIQFHE 209
Query: 751 STSCAP*RLNSRVPCTAY 698
+ A L VP T Y
Sbjct: 210 KSKPAKSTLFKVVPVTLY 227
>ref|ZP_00006349.1| COG1215: Glycosyltransferases, probably involved in cell wall
biogenesis [Rhodobacter sphaeroides]
Length = 788
Score = 33.9 bits (76), Expect = 4.3
Identities = 22/71 (30%), Positives = 33/71 (45%)
Frame = -1
Query: 794 RATAQTASGPTAPRLDKLCPVKAQLPGPVYGIYLASNLSAQQKVQGVRLTGVRLPKFAEH 615
RA A+ APR+ P +AQ+ P +G + GVRL VRLP +
Sbjct: 571 RAVAEKQQRRAAPRVQMEVPAEAQI--PAFGNRSLTATVLDASTSGVRLL-VRLPGVGDP 627
Query: 614 HPSQQCGAALK 582
HP+ + G ++
Sbjct: 628 HPALEAGGLIQ 638
>ref|XP_318131.1| ENSANGP00000017542 [Anopheles gambiae] gi|30175098|gb|EAA13166.2|
ENSANGP00000017542 [Anopheles gambiae str. PEST]
Length = 427
Score = 33.5 bits (75), Expect = 5.6
Identities = 25/78 (32%), Positives = 34/78 (43%)
Frame = -2
Query: 931 QLLRLVRRLSVCRKCACHTVHSIIQYLKHYRSTVSCTPTKEGPCCLEQLLKPLQVPRHHV 752
Q L+ V RL +CRKC + HSI Q YR ++ + L Q+ P + + H
Sbjct: 152 QRLQTVERLKLCRKCLNN--HSIEQCKTRYRCYINGCHKDDHHTLLHQVDVPQAIIQFHE 209
Query: 751 STSCAP*RLNSRVPCTAY 698
A L VP T Y
Sbjct: 210 KNKPAKSTLFKVVPVTLY 227
>ref|XP_317932.1| ENSANGP00000011364 [Anopheles gambiae] gi|30175206|gb|EAA12984.2|
ENSANGP00000011364 [Anopheles gambiae str. PEST]
Length = 427
Score = 33.5 bits (75), Expect = 5.6
Identities = 25/78 (32%), Positives = 34/78 (43%)
Frame = -2
Query: 931 QLLRLVRRLSVCRKCACHTVHSIIQYLKHYRSTVSCTPTKEGPCCLEQLLKPLQVPRHHV 752
Q L+ V RL +CRKC + HSI Q YR ++ + L Q+ P + + H
Sbjct: 152 QRLQTVERLKLCRKCLNN--HSIEQCKTRYRCYINGCHKDDHHTLLHQVDVPQAIIQFHE 209
Query: 751 STSCAP*RLNSRVPCTAY 698
A L VP T Y
Sbjct: 210 KNKPAKSTLFKVVPVTLY 227