Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000734A_C01 KCC000734A_c01
(1060 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
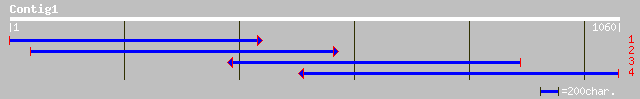
pir||T07921 probable starch synthase (EC 2.4.1.21), granule boun... 56 1e-06
gb|AAC17969.3| granule-bound starch synthase I precursor [Chlamy... 56 1e-06
gb|AAL31655.1|AC079179_10 Unknown protein [Oryza sativa] gi|2004... 52 2e-05
ref|NP_041033.1| tegument protein [Equine herpesvirus 1] gi|1355... 48 2e-04
dbj|BAB16871.1| putative protein kinase APK1AArabidopsis thalian... 48 3e-04
>pir||T07921 probable starch synthase (EC 2.4.1.21), granule bound - Chlamydomonas
reinhardtii (fragment)
Length = 238
Score = 55.8 bits (133), Expect = 1e-06
Identities = 28/28 (100%), Positives = 28/28 (100%)
Frame = -2
Query: 1059 SNGNGNGASASKTSAAKPLVSAATRKSA 976
SNGNGNGASASKTSAAKPLVSAATRKSA
Sbjct: 211 SNGNGNGASASKTSAAKPLVSAATRKSA 238
>gb|AAC17969.3| granule-bound starch synthase I precursor [Chlamydomonas reinhardtii]
gi|16755883|gb|AAL28128.1|AF433156_1 granule-bound starch
synthase I [Chlamydomonas reinhardtii]
Length = 708
Score = 55.8 bits (133), Expect = 1e-06
Identities = 28/28 (100%), Positives = 28/28 (100%)
Frame = -2
Query: 1059 SNGNGNGASASKTSAAKPLVSAATRKSA 976
SNGNGNGASASKTSAAKPLVSAATRKSA
Sbjct: 681 SNGNGNGASASKTSAAKPLVSAATRKSA 708
>gb|AAL31655.1|AC079179_10 Unknown protein [Oryza sativa] gi|20042881|gb|AAM08709.1|AC116601_2
Unknown protein [Oryza sativa] gi|31429914|gb|AAP51898.1|
unknown protein [Oryza sativa (japonica cultivar-group)]
Length = 1269
Score = 52.0 bits (123), Expect = 2e-05
Identities = 48/167 (28%), Positives = 61/167 (35%), Gaps = 11/167 (6%)
Frame = +3
Query: 195 PPRSTSTNPRFEPASKNRWPISQ*P*RAPKMSPRHPGLARALTPTVPSAPQQNHRG-ATQ 371
PPRS+S P S P P P PG+ A P P P G +
Sbjct: 671 PPRSSSRTPTGAATSSKGPPPPPPPPLPPANRTNGPGVPSAPPPPPPPPPANRSNGPSAP 730
Query: 372 APPLLPPF-------HTLLCIPSSTLGGHRATAPPTSPLPEAVGEPGAAVPPLTSHKMAN 530
APPL PP + P + G +A APP P P+A PG PP H +
Sbjct: 731 APPLPPPLPAAANKRNPPAPPPPPLMTGKKAPAPPPPP-PQAPKPPGTVPPPPPLHGASG 789
Query: 531 SQAACKNPGMDGVYRPALLPPNHKAEGRT---GCARAGVVAAPRKCS 662
+ G++ P LL +A G G A P+K S
Sbjct: 790 RPHPPSSKGLNAPAPPPLLGRGREATGSAKGRGIGLAQQSNPPKKAS 836
Score = 33.9 bits (76), Expect = 4.9
Identities = 16/34 (47%), Positives = 18/34 (52%), Gaps = 6/34 (17%)
Frame = -3
Query: 1058 PTATATVPRPPRPRLPSP------WSPPPPASPP 975
P+ TA P PP P P P +SPPPP PP
Sbjct: 537 PSPTAAAPPPPPPPPPPPSGNKPAFSPPPPPPPP 570
>ref|NP_041033.1| tegument protein [Equine herpesvirus 1]
gi|135577|sp|P28955|TEGU_HSVEB LARGE TEGUMENT PROTEIN
gi|73852|pir||WZBEB6 367K tegument protein - equine
herpesvirus 1 (strain Ab4p) gi|330816|gb|AAB02459.1|
tegument protein
Length = 3421
Score = 48.1 bits (113), Expect = 2e-04
Identities = 57/224 (25%), Positives = 88/224 (38%), Gaps = 8/224 (3%)
Frame = +3
Query: 114 NSPPMPPCRRTLHLVYLQS---SMSASFSNPPRSTSTNPRFEPASKNRWPISQ*-P*RAP 281
N+ P PP +T +V + S+ + + PP++ P A + P+ + P
Sbjct: 2480 NAMPAPP--KTTPVVESKQKSDSVDRAPTLPPKAAPLPPSDASAIMSGKPVFKYTPGNKS 2537
Query: 282 KMSPRHPGLARALTPTVPSAPQQNHRGATQAPPLLPPFHTLLCIPSSTLGGHRATAPPTS 461
+ P P A P P PQ + A+ PP LPP L S G T PP
Sbjct: 2538 AVPPSVP--APPTLPPAPPLPQSTSKAASGPPPTLPPAPPLPQSTSKAASGPPPTLPPAP 2595
Query: 462 PLPE----AVGEPGAAVPPLTSHKMANSQAACKNPGMDGVYRPALLPPNHKAEGRTGCAR 629
PLP+ A P +PP + S+AA D L P +++ +
Sbjct: 2596 PLPQSTSKAASGPPPTLPPAPPLPQSTSKAASGATQSDSGKTLTLDVPKTQSKDKV---- 2651
Query: 630 AGVVAAPRKCSATATVSLLFFWTWAKPKPQALHYKTKTGVSPVS 761
V P +T T + L +KP A+ ++ K G +PV+
Sbjct: 2652 --VPVPPTDKPSTTTPAALKQSDASKPPTAAIQHQQKLG-TPVT 2692
>dbj|BAB16871.1| putative protein kinase APK1AArabidopsis thaliana [Oryza sativa
(japonica cultivar-group)] gi|14646774|dbj|BAB61937.1|
putative protein kinase APK1AArabidopsis thaliana [Oryza
sativa (japonica cultivar-group)]
Length = 1014
Score = 47.8 bits (112), Expect = 3e-04
Identities = 58/196 (29%), Positives = 81/196 (40%), Gaps = 3/196 (1%)
Frame = +3
Query: 195 PPRSTSTNPR-FEPASKNRWPISQ*P*RAPKMSPRHPGLARALTPTVPSAPQQNHRGATQ 371
PP++ NP P + P Q P AP ++ R+P L P+V + P +
Sbjct: 114 PPQAAVENPAPVLPGTPALLPSVQAP--APSVA-RNPNLPIVQPPSVNNPPSRPIGSGNG 170
Query: 372 APPLLPPFHTLLCIPSSTLGGHR-ATAPPTSPLPEAVGEPGAAVPPLTSHKMANS-QAAC 545
PP PP +L IP ST G R + PP +P A A+ P + H NS A
Sbjct: 171 VPPYPPPQRSLPAIPPSTSGVPRESVKPPVAPPIIAQAPRQQALAPSSDHSNGNSVPPAN 230
Query: 546 KNPGMDGVYRPALLPPNHKAEGRTGCARAGVVAAPRKCSATATVSLLFFWTWAKPKPQAL 725
+P + P LPP ++ +TG A P + SA A L P+ +
Sbjct: 231 TSPPHKNSHIPRALPPK-ESSSQTGTAH----KPPIRGSAPAETPL--------PQNTNM 277
Query: 726 HYKTKTGVSPVSQNRP 773
K G S VS +RP
Sbjct: 278 PAVPKNG-SSVSHDRP 292
Score = 33.9 bits (76), Expect = 4.9
Identities = 34/104 (32%), Positives = 41/104 (38%), Gaps = 3/104 (2%)
Frame = +3
Query: 288 SPRHPGLARALTP-TVPSAPQQNHRGATQAPPLLPPFHTLLCIPSSTLGGHRATAPPTSP 464
SP P +A L P T+P+AP AP PP P+ A P T P
Sbjct: 41 SPPEPTIA--LGPVTLPTAPS--------APSASPPVAKGAVSPAVPTRPQNAPTPVTPP 90
Query: 465 LPEAVGEPGAAVPPLTSH--KMANSQAACKNPGMDGVYRPALLP 590
P PP +H +A QAA +NP PALLP
Sbjct: 91 KEYNAPPPVELTPPAPTHVAPVAPPQAAVENPAPVLPGTPALLP 134