Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000698A_C01 KCC000698A_c01
(570 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
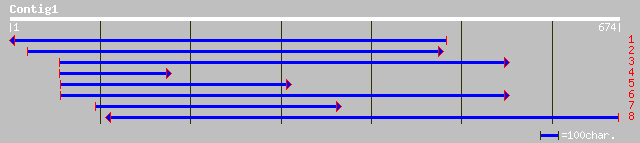
ref|NP_178688.1| plant transposase (Ptta/En/Spm) family [Arabido... 37 0.15
ref|ZP_00129015.1| COG2204: Response regulator containing CheY-l... 34 1.6
dbj|BAC85954.1| unnamed protein product [Homo sapiens] 28 2.1
gb|AAO51593.1| similar to Plasmodium falciparum (isolate 3D7). H... 33 3.6
ref|ZP_00019363.1| hypothetical protein [Chloroflexus aurantiacus] 32 6.1
>ref|NP_178688.1| plant transposase (Ptta/En/Spm) family [Arabidopsis thaliana]
gi|25411243|pir||H84477 probable PttA-like transposon
protein [imported] - Arabidopsis thaliana
gi|4584354|gb|AAD25148.1| putative PttA-like transposon
protein [Arabidopsis thaliana]
Length = 491
Score = 37.4 bits (85), Expect = 0.15
Identities = 28/86 (32%), Positives = 37/86 (42%), Gaps = 4/86 (4%)
Frame = +1
Query: 178 GHDGSQAAGTHNSATPHLLHRCYMTSYLPATVPQLTPYTHVVMCRNVIRPVSPAQSYNRT 357
GH S + NSAT HR + Y PATVPQ TP + N +P+
Sbjct: 43 GHRASSHSSNPNSATQS--HRSRPSQYPPATVPQTTPPQTTLTPTNSRQPLLSGSQQPPP 100
Query: 358 HTQSQSPNR----PSDQQSPCPQEQK 423
++ P R P+ QQ P +QK
Sbjct: 101 VHRNLPPTRQRQAPTSQQQPPTCQQK 126
>ref|ZP_00129015.1| COG2204: Response regulator containing CheY-like receiver, AAA-type
ATPase, and DNA-binding domains [Desulfovibrio
desulfuricans G20]
Length = 586
Score = 33.9 bits (76), Expect = 1.6
Identities = 26/68 (38%), Positives = 32/68 (46%), Gaps = 6/68 (8%)
Frame = +3
Query: 156 HVRTYTSRARRQSGRRNSQFRDSTSAAPVLHD*LSTCDGAPTN------PIHTCGHVP*C 317
HVR T+RA Q R DST+AA D T GAP + P HTCG +P
Sbjct: 433 HVRISTARAALQQRRT-----DSTAAAQPSRD-EDTASGAPESRPQAPRPCHTCGTLPES 486
Query: 318 YPSCVPRP 341
Y + +P
Sbjct: 487 YGTPAVQP 494
>dbj|BAC85954.1| unnamed protein product [Homo sapiens]
Length = 189
Score = 28.5 bits (62), Expect(2) = 2.1
Identities = 11/27 (40%), Positives = 16/27 (58%), Gaps = 1/27 (3%)
Frame = +1
Query: 97 VHFTCYKYRL-LCKQACLGVCMCAHTH 174
+H C + +C AC+ VC CA+TH
Sbjct: 77 IHIMCMHVCVYVCVHACMCVCTCAYTH 103
Score = 23.9 bits (50), Expect(2) = 2.1
Identities = 16/61 (26%), Positives = 22/61 (35%)
Frame = +2
Query: 149 VCACAHIHITGTTAVRPQELTIPRLHICCTGAT*LAIYLRRCPN*PHTHMWSCAVMLSVL 328
VC CA+ H+ VR HIC +Y C HM C + + +
Sbjct: 96 VCTCAYTHVFVCVRVRTCVYMRTHTHIC--------VYACAC-----AHMCVCVRVHTYV 142
Query: 329 C 331
C
Sbjct: 143 C 143
>gb|AAO51593.1| similar to Plasmodium falciparum (isolate 3D7). Hypothetical
protein [Dictyostelium discoideum]
Length = 1845
Score = 32.7 bits (73), Expect = 3.6
Identities = 22/68 (32%), Positives = 30/68 (43%)
Frame = +1
Query: 340 QSYNRTHTQSQSPNRPSDQQSPCPQEQK*RNKRPVKWSCGLFLKRPYVPPPICSNAVSIP 519
QS +Q Q P +P +QQ P P RN+ P K KRP P S P
Sbjct: 141 QSQQSQQSQQQQPQQPQEQQEPLPNLSFLRNQEPDKNVLPTSRKRPPSVMP------STP 194
Query: 520 HQTTTNFS 543
+Q++ + S
Sbjct: 195 NQSSNSSS 202
>ref|ZP_00019363.1| hypothetical protein [Chloroflexus aurantiacus]
Length = 354
Score = 32.0 bits (71), Expect = 6.1
Identities = 21/68 (30%), Positives = 28/68 (40%), Gaps = 2/68 (2%)
Frame = +3
Query: 255 LSTCDGAPTNPIHTCGHVP*CYPSCVPRPVI*PY--THSVSEPQPTK*PAVTLPPGTKIE 428
LS C T + T P P+ +P P P T V+EP+PT + P T +
Sbjct: 18 LSACGAGSTATVPTAVPEPTAIPTAIPAPTAAPVPTTAPVAEPEPTSAVLTPVLPATVTD 77
Query: 429 KQATREVV 452
Q VV
Sbjct: 78 YQGETVVV 85