Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000666A_C01 KCC000666A_c01
(523 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
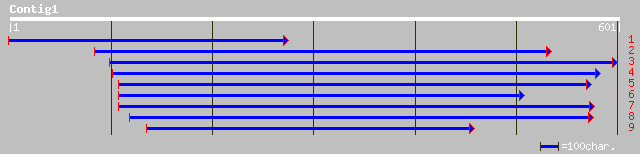
gb|AAK30605.1|AF350276_1 major ampullate spidroin 2-like protein... 41 0.011
ref|NP_627816.1| putative serine-threonine protein kinase [Strep... 41 0.011
ref|NP_857510.1| PROBABLE CONSERVED TRANSMEMBRANE PROTEIN [Mycob... 40 0.014
ref|XP_313643.1| ENSANGP00000013121 [Anopheles gambiae] gi|21296... 40 0.018
sp|P46804|SPD2_NEPCL Spidroin 2 (Dragline silk fibroin 2) gi|345... 40 0.018
>gb|AAK30605.1|AF350276_1 major ampullate spidroin 2-like protein [Nephila madagascariensis]
Length = 1953
Score = 40.8 bits (94), Expect = 0.011
Identities = 34/87 (39%), Positives = 37/87 (42%), Gaps = 9/87 (10%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG G A G GP G QG +GP PG +A A Q G A A G
Sbjct: 732 GGPGAAAAAA-AGRGPGGYGQGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAGSG 789
Query: 312 PGGGERSRSWPGSSQAAAPARPAEPGP 392
PGG + PG S AAA A A GP
Sbjct: 790 PGGYGPGQQGPGRSGAAAAAAAAGRGP 816
Score = 40.8 bits (94), Expect = 0.011
Identities = 34/87 (39%), Positives = 37/87 (42%), Gaps = 9/87 (10%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG G A G GP G QG +GP PG +A A Q G A A G
Sbjct: 1348 GGPGAAAAAA-AGRGPGGYGQGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAGSG 1405
Query: 312 PGGGERSRSWPGSSQAAAPARPAEPGP 392
PGG + PG S AAA A A GP
Sbjct: 1406 PGGYGPGQQGPGRSGAAAAAAAAGRGP 1432
Score = 38.5 bits (88), Expect = 0.053
Identities = 31/86 (36%), Positives = 35/86 (40%), Gaps = 9/86 (10%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
G +G A G GP G G +GP PG +A A Q G A A G
Sbjct: 1417 GRSGAAAAAAAAGRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAASAGRG 1475
Query: 312 PGGGERSRSWPGSSQAAAPARPAEPG 389
PGG + PG S AAA A PG
Sbjct: 1476 PGGYGPGQQGPGGSGAAAAAAGRGPG 1501
Score = 38.5 bits (88), Expect = 0.053
Identities = 28/83 (33%), Positives = 33/83 (39%), Gaps = 11/83 (13%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPD--------VLPGDSALAQNGAFAT*EAWLGGWP 314
GG G A G GP G G +GP PG Q G A A G P
Sbjct: 1278 GGPGAAAAAAAAGRGPGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGPGAAAAAAAGSGP 1337
Query: 315 GG---GERSRSWPGSSQAAAPAR 374
GG G++ PG++ AAA R
Sbjct: 1338 GGYGPGQQGPGGPGAAAAAAAGR 1360
Score = 38.5 bits (88), Expect = 0.053
Identities = 31/86 (36%), Positives = 35/86 (40%), Gaps = 9/86 (10%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
G +G A G GP G G +GP PG +A A Q G A A G
Sbjct: 801 GRSGAAAAAAAAGRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAASAGRG 859
Query: 312 PGGGERSRSWPGSSQAAAPARPAEPG 389
PGG + PG S AAA A PG
Sbjct: 860 PGGYGPGQQGPGGSGAAAAAAGRGPG 885
Score = 37.0 bits (84), Expect = 0.15
Identities = 32/90 (35%), Positives = 39/90 (42%), Gaps = 12/90 (13%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG+G A G GP G G +GP PG +A A Q G A A G
Sbjct: 925 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAGSG 981
Query: 312 PGG---GERSRSWPGSSQAAAPARPAEPGP 392
PGG G++ PG++ AAA P GP
Sbjct: 982 PGGYGPGQQGPGGPGAAAAAAGRGPGGYGP 1011
Score = 37.0 bits (84), Expect = 0.15
Identities = 32/90 (35%), Positives = 39/90 (42%), Gaps = 12/90 (13%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG+G A G GP G G +GP PG +A A Q G A A G
Sbjct: 1536 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAGSG 1592
Query: 312 PGG---GERSRSWPGSSQAAAPARPAEPGP 392
PGG G++ PG++ AAA P GP
Sbjct: 1593 PGGYGPGQQGPGGPGAAAAAAGRGPGGYGP 1622
Score = 35.8 bits (81), Expect = 0.34
Identities = 31/80 (38%), Positives = 34/80 (41%), Gaps = 3/80 (3%)
Frame = +3
Query: 162 GTGEATKGTKCGVGPSGVSQGCRGPDVL---PGDSALAQNGAFAT*EAWLGGWPGGGERS 332
G A G++ G GP QG RGP PG Q GA A A G PGG
Sbjct: 136 GAAAAAAGSE-GYGPG--QQGPRGPGAAAAGPGGYGPGQQGASAAASAAAGRGPGGYGPG 192
Query: 333 RSWPGSSQAAAPARPAEPGP 392
+ PG AAA A P GP
Sbjct: 193 QQGPGGPSAAA-AGPGGYGP 211
Score = 35.4 bits (80), Expect = 0.45
Identities = 30/92 (32%), Positives = 33/92 (35%), Gaps = 14/92 (15%)
Frame = +3
Query: 159 GGTGEATKG------TKCGVGPSGVSQGCRGPD--------VLPGDSALAQNGAFAT*EA 296
GG G +G G GP G G +GP PG Q G A A
Sbjct: 1202 GGYGPGQQGPGAAAAAAAGSGPGGYGPGQQGPGGSSAAAAAAGPGRYGPGQQGPGAAAAA 1261
Query: 297 WLGGWPGGGERSRSWPGSSQAAAPARPAEPGP 392
G PGG + PG AAA A A GP
Sbjct: 1262 AAGSGPGGYGPGQQGPGGPGAAAAAAAAGRGP 1293
Score = 35.0 bits (79), Expect = 0.59
Identities = 27/76 (35%), Positives = 30/76 (38%)
Frame = +3
Query: 162 GTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRSW 341
G G A G GP G G +GP PG +A A G PGG +
Sbjct: 1697 GPGAAAAAAAAGRGPGGYGPGQQGPGG-PGAAAAAAAGR----------GPGGYGPGQQG 1745
Query: 342 PGSSQAAAPARPAEPG 389
PG S AAA A PG
Sbjct: 1746 PGGSGAAAAAAGRGPG 1761
Score = 34.7 bits (78), Expect = 0.77
Identities = 33/94 (35%), Positives = 37/94 (39%), Gaps = 16/94 (17%)
Frame = +3
Query: 159 GGTGEATKGT------KCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*E 293
GG G +GT G G G G +GP PG +A A Q G A
Sbjct: 1158 GGYGPGQQGTGAAAAAAAGSGAGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAA 1216
Query: 294 AWLGGWPGGGERSRSWPGSSQAAAPAR-PAEPGP 392
A G PGG + PG S AAA A P GP
Sbjct: 1217 AAAGSGPGGYGPGQQGPGGSSAAAAAAGPGRYGP 1250
Score = 34.7 bits (78), Expect = 0.77
Identities = 33/94 (35%), Positives = 37/94 (39%), Gaps = 16/94 (17%)
Frame = +3
Query: 159 GGTGEATKGT------KCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*E 293
GG G +GT G G G G +GP PG +A A Q G A
Sbjct: 568 GGYGPGQQGTGAAAAAAAGSGAGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAA 626
Query: 294 AWLGGWPGGGERSRSWPGSSQAAAPAR-PAEPGP 392
A G PGG + PG S AAA A P GP
Sbjct: 627 AAAGSGPGGYGPGQQGPGGSSAAAAAAGPGRYGP 660
Score = 34.3 bits (77), Expect = 1.0
Identities = 31/87 (35%), Positives = 35/87 (39%), Gaps = 10/87 (11%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA----------QNGAFAT*EAWLGG 308
GG+G A G GP G G +GP PG +A A Q A A G
Sbjct: 463 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAAGR 519
Query: 309 WPGGGERSRSWPGSSQAAAPARPAEPG 389
PGG + PG S AAA A PG
Sbjct: 520 GPGGYGPGQQGPGGSGAAAAAAGRGPG 546
Score = 33.9 bits (76), Expect = 1.3
Identities = 30/96 (31%), Positives = 34/96 (35%), Gaps = 18/96 (18%)
Frame = +3
Query: 159 GGTGEATKGTKC------GVGPSGVSQGCRGPD------VLPGDSALAQNGAFAT*EAWL 302
GG G +G G GP G G +GP PG Q G A A
Sbjct: 165 GGYGPGQQGASAAASAAAGRGPGGYGPGQQGPGGPSAAAAGPGGYGPGQQGPSAAAAAAA 224
Query: 303 GGWPGGGERSRSWPGSSQAAAPAR------PAEPGP 392
G PGG + PG AAA A P + GP
Sbjct: 225 GSGPGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGP 260
Score = 33.9 bits (76), Expect = 1.3
Identities = 30/85 (35%), Positives = 33/85 (38%), Gaps = 8/85 (9%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPD--------VLPGDSALAQNGAFAT*EAWLGGWP 314
GG+G A G GP G G +GP PG Q G A A G P
Sbjct: 1747 GGSGAAAAAA--GRGPGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGPGAA-AAAAGRGP 1803
Query: 315 GGGERSRSWPGSSQAAAPARPAEPG 389
GG + PG S AAA A PG
Sbjct: 1804 GGYGPGQQGPGGSGAAAAAAGRGPG 1828
Score = 33.9 bits (76), Expect = 1.3
Identities = 28/77 (36%), Positives = 32/77 (41%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRS 338
GG+G A G GP G G +GP PG +A A G PGG +
Sbjct: 1487 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAAGT----------GPGGYGPGQQ 1533
Query: 339 WPGSSQAAAPARPAEPG 389
PG S AAA A PG
Sbjct: 1534 GPGGSGAAAAAAGRGPG 1550
Score = 32.7 bits (73), Expect = 2.9
Identities = 32/97 (32%), Positives = 40/97 (40%), Gaps = 19/97 (19%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA----------QNGAFAT*EAWLGG 308
GG+G A G GP G G +GP PG +A A Q A A G
Sbjct: 1047 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAAGR 1103
Query: 309 WPGG---GERSRSWPGSSQAAAPAR------PAEPGP 392
PGG G++ PG++ AAA R P + GP
Sbjct: 1104 GPGGYGPGQQGPGGPGAAAAAAAGRGPGGYGPGQQGP 1140
Score = 32.7 bits (73), Expect = 2.9
Identities = 28/77 (36%), Positives = 31/77 (39%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRS 338
GG G A G GP G G +GP PG +A A G PGG +
Sbjct: 1604 GGPGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAAGR----------GPGGYGPGQQ 1650
Query: 339 WPGSSQAAAPARPAEPG 389
PG S AAA A PG
Sbjct: 1651 GPGGSGAAAAASGRGPG 1667
Score = 32.7 bits (73), Expect = 2.9
Identities = 26/90 (28%), Positives = 32/90 (34%), Gaps = 12/90 (13%)
Frame = +3
Query: 159 GGTGEATKG----TKCGVGPSGVSQGCRGPDVL--------PGDSALAQNGAFAT*EAWL 302
GG G +G + GP G G +GP PG Q G A
Sbjct: 187 GGYGPGQQGPGGPSAAAAGPGGYGPGQQGPSAAAAAAAGSGPGGYGPGQQGPGGPGAAAA 246
Query: 303 GGWPGGGERSRSWPGSSQAAAPARPAEPGP 392
PGG + PG++ AAA P GP
Sbjct: 247 AAGPGGYGPGQQGPGAAAAAAGRGPGGYGP 276
Score = 32.0 bits (71), Expect = 5.0
Identities = 26/77 (33%), Positives = 32/77 (40%)
Frame = +3
Query: 162 GTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRSW 341
G G A G GP G G +GP +A A G GG+ G G++
Sbjct: 507 GPGAAAAAAAAGRGPGGYGPGQQGPGGSGAAAAAAGRGP--------GGY-GPGQQGPGG 557
Query: 342 PGSSQAAAPARPAEPGP 392
PG+ AAA A P GP
Sbjct: 558 PGA--AAAAAGPGGYGP 572
Score = 31.6 bits (70), Expect = 6.5
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 3/81 (3%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGG---GER 329
GG G A G GP G G +GP G +GA A A G PGG G++
Sbjct: 334 GGPGAAAAAA-AGRGPGGYGPGQQGP----GQQGPGGSGAAA---AAAGRGPGGYGPGQQ 385
Query: 330 SRSWPGSSQAAAPARPAEPGP 392
PG+ AAA A P GP
Sbjct: 386 GPGGPGA--AAAAAGPGGYGP 404
Score = 31.6 bits (70), Expect = 6.5
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 3/81 (3%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGG---GER 329
GG G A G GP G G +GP G +GA A A G PGG G++
Sbjct: 1017 GGPGAAAAAA-AGRGPGGYGPGQQGP----GQQGPGGSGAAA---AAAGRGPGGYGPGQQ 1068
Query: 330 SRSWPGSSQAAAPARPAEPGP 392
PG+ AAA A P GP
Sbjct: 1069 GPGGPGA--AAAAAGPGGYGP 1087
Score = 31.6 bits (70), Expect = 6.5
Identities = 31/93 (33%), Positives = 36/93 (38%), Gaps = 15/93 (16%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG+G A G GP G G +GP PG +A A Q G A A G
Sbjct: 1814 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGTGAAAAAAAGSG 1870
Query: 312 PGGGERSRSWPGSSQAAAPAR------PAEPGP 392
GG + PG AAA A P + GP
Sbjct: 1871 AGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGP 1903
Score = 31.6 bits (70), Expect = 6.5
Identities = 31/93 (33%), Positives = 36/93 (38%), Gaps = 15/93 (16%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGW 311
GG+G A G GP G G +GP PG +A A Q G A A G
Sbjct: 532 GGSGAAAAAA--GRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGTGAAAAAAAGSG 588
Query: 312 PGGGERSRSWPGSSQAAAPAR------PAEPGP 392
GG + PG AAA A P + GP
Sbjct: 589 AGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGP 621
Score = 31.6 bits (70), Expect = 6.5
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 3/81 (3%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGG---GER 329
GG G A G GP G G +GP G +GA A A G PGG G++
Sbjct: 895 GGPGAAAAAA-AGRGPGGYGPGQQGP----GQQGPGGSGAAA---AAAGRGPGGYGPGQQ 946
Query: 330 SRSWPGSSQAAAPARPAEPGP 392
PG+ AAA A P GP
Sbjct: 947 GPGGPGA--AAAAAGPGGYGP 965
Score = 31.6 bits (70), Expect = 6.5
Identities = 30/98 (30%), Positives = 34/98 (34%), Gaps = 20/98 (20%)
Frame = +3
Query: 159 GGTGEATKG------TKCGVGPSGVSQGCRGPD--------VLPGDSALAQNGAFAT*EA 296
GG G +G G GP G G +GP PG Q G A A
Sbjct: 612 GGYGPGQQGPGAAAAAAAGSGPGGYGPGQQGPGGSSAAAAAAGPGRYGPGQQGPGAAAAA 671
Query: 297 WLGGWPGGGERSRSWPGSSQAAAPAR------PAEPGP 392
G PGG + PG AAA A P + GP
Sbjct: 672 SAGRGPGGYGPGQQGPGGPGAAAAAAGPGGYGPGQQGP 709
Score = 31.6 bits (70), Expect = 6.5
Identities = 30/81 (37%), Positives = 35/81 (43%), Gaps = 3/81 (3%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGG---GER 329
GG G A G GP G G +GP G +GA A A G PGG G++
Sbjct: 433 GGPGAAAAAA-AGRGPGGYGPGQQGP----GQQGPGGSGAAA---AAAGRGPGGYGPGQQ 484
Query: 330 SRSWPGSSQAAAPARPAEPGP 392
PG+ AAA A P GP
Sbjct: 485 GPGGPGA--AAAAAGPGGYGP 503
Score = 31.2 bits (69), Expect = 8.5
Identities = 26/75 (34%), Positives = 29/75 (38%), Gaps = 9/75 (12%)
Frame = +3
Query: 174 ATKGTKCGVGPSGVSQGCRGPDVLPGDSALA---------QNGAFAT*EAWLGGWPGGGE 326
A G GP G G +GP PG +A A Q G A A G PGG
Sbjct: 667 AAAAASAGRGPGGYGPGQQGPGG-PGAAAAAAGPGGYGPGQQGPGAAAAAAAGSGPGGYG 725
Query: 327 RSRSWPGSSQAAAPA 371
+ PG AAA A
Sbjct: 726 PGQQGPGGPGAAAAA 740
>ref|NP_627816.1| putative serine-threonine protein kinase [Streptomyces coelicolor
A3(2)] gi|7481393|pir||T35389 probable serine-threonine
protein kinase - Streptomyces coelicolor
gi|5123896|emb|CAB45488.1| putative serine-threonine
protein kinase [Streptomyces coelicolor A3(2)]
Length = 783
Score = 40.8 bits (94), Expect = 0.011
Identities = 29/78 (37%), Positives = 32/78 (40%)
Frame = +3
Query: 159 GGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRS 338
GG G G+ GVGP G G GP G + +GA G WPGGG
Sbjct: 427 GGVGPGGAGSG-GVGPGGAGSGGVGPGGA-GSGSAGPDGADPGGVGPGGAWPGGGGARGG 484
Query: 339 WPGSSQAAAPARPAEPGP 392
G AA A PA GP
Sbjct: 485 GSGGEGAAQGADPASGGP 502
>ref|NP_857510.1| PROBABLE CONSERVED TRANSMEMBRANE PROTEIN [Mycobacterium bovis
subsp. bovis AF2122/97] gi|31620615|emb|CAD96059.1|
PROBABLE CONSERVED TRANSMEMBRANE PROTEIN [Mycobacterium
bovis subsp. bovis AF2122/97]
Length = 342
Score = 40.4 bits (93), Expect = 0.014
Identities = 32/96 (33%), Positives = 45/96 (46%), Gaps = 7/96 (7%)
Frame = -1
Query: 457 CCSRQDLRQLQRCWCPWAQRRTGPGSAGLAGAAACELPGQLLLRSPPPGQPPSQASHVAK 278
C +R ++R+ QR WCP + G+ A ++P + R+P Q P+ AS +
Sbjct: 8 CGTRWNVRERQRVWCPRCR--------GMLLAPLADMPAEARWRTPARPQVPT-ASDTRR 58
Query: 277 AP------F*ARALSPGNTSGPRH-PWLTPLGPTPH 191
P F A+ PG PRH P L GPTPH
Sbjct: 59 TPPRLPPGFRWIAVRPGAAPPPRHGPRLR--GPTPH 92
>ref|XP_313643.1| ENSANGP00000013121 [Anopheles gambiae] gi|21296935|gb|EAA09080.1|
ENSANGP00000013121 [Anopheles gambiae str. PEST]
Length = 396
Score = 40.0 bits (92), Expect = 0.018
Identities = 32/100 (32%), Positives = 45/100 (45%), Gaps = 11/100 (11%)
Frame = -3
Query: 398 KNRSWFCGSCRSRSLRAARPAPAALPTP------RPAAEPSFSRSKSSILSKSTVSR*YI 237
+ +W+C CRS+ A PA +PTP P+ S SKSS SKS S
Sbjct: 209 EENAWYCTICRSKQAVAKLATPAVVPTPPIPVATSPSPTIGSSSSKSSSHSKSGSSSKSA 268
Query: 236 RSSASLADSAWADTTFSSFCSFSCSSMYQ-----DRSRSE 132
SS+S + S+ + S + S S ++ DR R E
Sbjct: 269 ESSSSSSSSSSSKHKSSKSGNKSSDSSHERDRDRDRDRGE 308
>sp|P46804|SPD2_NEPCL Spidroin 2 (Dragline silk fibroin 2) gi|345426|pir||A44112 spidroin
2, dragline silk fibroin - orb spider (Nephila clavipes)
(fragment) gi|159714|gb|AAA29381.1| dragline silk
fibroin
Length = 627
Score = 40.0 bits (92), Expect = 0.018
Identities = 28/73 (38%), Positives = 36/73 (48%), Gaps = 9/73 (12%)
Frame = +3
Query: 201 GPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGW------PGG---GERSRSWPGSS 353
GP G G +GP PG +A A A + GG+ PGG G++ S PGS+
Sbjct: 188 GPGGYGPGQQGPSG-PGSAAAAAAAASGPGQQGPGGYGPGQQGPGGYGPGQQGLSGPGSA 246
Query: 354 QAAAPARPAEPGP 392
AAA A P + GP
Sbjct: 247 AAAAAAGPGQQGP 259
Score = 37.4 bits (85), Expect = 0.12
Identities = 25/63 (39%), Positives = 31/63 (48%)
Frame = +3
Query: 201 GPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGWPGGGERSRSWPGSSQAAAPARPA 380
GP G G +GP SA A A + GG+ G G++ S PGS+ AAA A A
Sbjct: 333 GPGGYGPGQQGPGGYGPGSASAAAAAAGPGQQGPGGY-GPGQQGPSGPGSASAAAAAAAA 391
Query: 381 EPG 389
PG
Sbjct: 392 GPG 394
Score = 35.0 bits (79), Expect = 0.59
Identities = 27/68 (39%), Positives = 34/68 (49%), Gaps = 5/68 (7%)
Frame = +3
Query: 201 GPSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGW--PGG---GERSRSWPGSSQAAA 365
GP G G +GP PG ++ A A A + G PGG G++ S PGS+ AAA
Sbjct: 365 GPGGYGPGQQGPSG-PGSASAAAAAAAAGPGGYGPGQQGPGGYAPGQQGPSGPGSASAAA 423
Query: 366 PARPAEPG 389
A A PG
Sbjct: 424 AAAAAGPG 431
Score = 33.5 bits (75), Expect = 1.7
Identities = 35/105 (33%), Positives = 45/105 (42%), Gaps = 12/105 (11%)
Frame = +3
Query: 114 PVEPSYLRA*AVLVHGGTGEATKGTKCGVGPSGVSQGCRGPDVLPGDSALAQNGAFAT*E 293
P P A A G G G + GP G + G +GP PG ++ A A A
Sbjct: 376 PSGPGSASAAAAAAAAGPGGYGPGQQ---GPGGYAPGQQGPSG-PGSASAAAAAAAAGPG 431
Query: 294 AWLGGW--PGG---GERSRSWPGSSQAAAPAR-------PAEPGP 392
+ G PGG G++ S PGS+ AAA A PA+ GP
Sbjct: 432 GYGPGQQGPGGYAPGQQGPSGPGSAAAAAAAAAGPGGYGPAQQGP 476
Score = 32.3 bits (72), Expect = 3.8
Identities = 30/96 (31%), Positives = 37/96 (38%), Gaps = 19/96 (19%)
Frame = +3
Query: 162 GTGEATKGTKCGVGPSGVSQGCRGPDVL-PGDSALAQNGAFAT*EA------WLGGW--- 311
G G A GP G G +GP PG + G+ A A LGG+
Sbjct: 271 GPGSAAAAAAAAAGPGGYGPGQQGPGGYGPGQQGPSGAGSAAAAAAAGPGQQGLGGYGPG 330
Query: 312 ---PGGGERSRSWPG------SSQAAAPARPAEPGP 392
PGG + PG +S AAA A P + GP
Sbjct: 331 QQGPGGYGPGQQGPGGYGPGSASAAAAAAGPGQQGP 366
Score = 32.3 bits (72), Expect = 3.8
Identities = 29/86 (33%), Positives = 38/86 (43%), Gaps = 22/86 (25%)
Frame = +3
Query: 201 GPSGVSQGCRGPDVLPGDSALAQN-----------GAFAT*EAWLGGW------PGG--- 320
GP G QG +GP PG +A A G + + GG+ PGG
Sbjct: 95 GPGGYGQGQQGPSG-PGSAAAASAAASAESGQQGPGGYGPGQQGPGGYGPGQQGPGGYGP 153
Query: 321 GERSRSWPGSSQAAAPAR--PAEPGP 392
G++ S PGS+ AAA A P + GP
Sbjct: 154 GQQGPSGPGSAAAAAAAASGPGQQGP 179
Score = 32.0 bits (71), Expect = 5.0
Identities = 30/107 (28%), Positives = 43/107 (40%)
Frame = -1
Query: 520 RRGAANPAAHLSACPRGPGRTCCSRQDLRQLQRCWCPWAQRRTGPGSAGLAGAAACELPG 341
++G + P + +A GPG+ Q + P Q +GPGSA A AAA G
Sbjct: 237 QQGLSGPGSAAAAAAAGPGQ---------QGPGGYGPGQQGPSGPGSAAAAAAAAAGPGG 287
Query: 340 QLLLRSPPPGQPPSQASHVAKAPF*ARALSPGNTSGPRHPWLTPLGP 200
+ P G P Q + P A + + +GP L GP
Sbjct: 288 YGPGQQGPGGYGPGQ-----QGPSGAGSAAAAAAAGPGQQGLGGYGP 329
Score = 31.6 bits (70), Expect = 6.5
Identities = 39/126 (30%), Positives = 48/126 (37%), Gaps = 9/126 (7%)
Frame = +3
Query: 162 GTGEATKGTKCGVG---PSGVSQGCRGPDVLPGDSALAQNGAFAT*EAWLGGW------P 314
G G A G G P G G +GP PG +A A A A GG+ P
Sbjct: 242 GPGSAAAAAAAGPGQQGPGGYGPGQQGPSG-PGSAAAAAAAA-----AGPGGYGPGQQGP 295
Query: 315 GGGERSRSWPGSSQAAAPARPAEPGPVLL*AHGHQQRCSCRKSWREQQVLPGPRGQADR* 494
GG + P + +AA A A PG L +G Q+ + Q PG G
Sbjct: 296 GGYGPGQQGPSGAGSAAAAAAAGPGQQGLGGYGPGQQGP--GGYGPGQQGPGGYGPGSAS 353
Query: 495 AAGLAA 512
AA AA
Sbjct: 354 AAAAAA 359
Score = 31.2 bits (69), Expect = 8.5
Identities = 28/69 (40%), Positives = 34/69 (48%), Gaps = 6/69 (8%)
Frame = +3
Query: 201 GPSGVSQGCRGPDVL-PGDSALAQNGAFAT*EAWLGGW--PGG---GERSRSWPGSSQAA 362
GP G G +GP PG L+ G+ A A G PGG G++ S PGS+ AA
Sbjct: 219 GPGGYGPGQQGPGGYGPGQQGLSGPGSAAAAAAAGPGQQGPGGYGPGQQGPSGPGSAAAA 278
Query: 363 APARPAEPG 389
A A A PG
Sbjct: 279 A-AAAAGPG 286