Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000560A_C01 KCC000560A_c01
(517 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
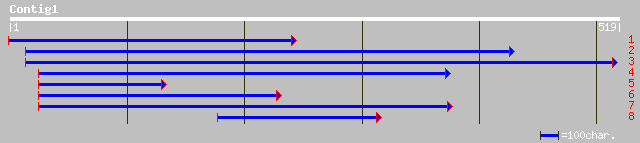
ref|XP_315792.1| ENSANGP00000018663 [Anopheles gambiae] gi|21299... 36 0.33
ref|NP_788733.1| multiple ankyrin repeats single KH domain CG331... 35 0.57
gb|AAL65911.1|AF425651_1 multiple ankyrin repeat single KH domai... 35 0.57
gb|AAL68383.1| SD05950p [Drosophila melanogaster] 35 0.57
gb|AAM11086.1| GH26759p [Drosophila melanogaster] 35 0.57
>ref|XP_315792.1| ENSANGP00000018663 [Anopheles gambiae] gi|21299754|gb|EAA11899.1|
ENSANGP00000018663 [Anopheles gambiae str. PEST]
Length = 944
Score = 35.8 bits (81), Expect = 0.33
Identities = 29/121 (23%), Positives = 51/121 (41%), Gaps = 1/121 (0%)
Frame = -1
Query: 505 HRRSTATLGTWRSLKRECCSTGSLDTAIETVPLLKLAAVQRSRPSAGTPPN-AGRGSAPR 329
H+R A G K TG++ ++ L +++ + P +G P+ GS P
Sbjct: 2 HKRHVAAAGRKSMEKFPLKGTGNISSSNN----LSISSAATNHPLSGDHPSYQSVGSKPG 57
Query: 328 RGSAALRYPRHFCSSLALGKPSPRGQSARAYAAKARGAGSLDLSQLQGSLQSPSNYSSSY 149
+GS A RH ++ + G +A G +D++ G L +P+ SSY
Sbjct: 58 KGSPASGAERHITTTTTTSLTTAPGTVGAVPVYEADEDGMIDITPGNGGLTNPNYSYSSY 117
Query: 148 T 146
+
Sbjct: 118 S 118
>ref|NP_788733.1| multiple ankyrin repeats single KH domain CG33106-PA [Drosophila
melanogaster] gi|28571867|ref|NP_788734.1| multiple
ankyrin repeats single KH domain CG33106-PB [Drosophila
melanogaster] gi|28381452|gb|AAO41600.1| CG33106-PA
[Drosophila melanogaster] gi|28381453|gb|AAO41601.1|
CG33106-PB [Drosophila melanogaster]
Length = 4001
Score = 35.0 bits (79), Expect = 0.57
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = -1
Query: 448 STGSLDTAIETVPLLKLAAVQRSRPSAGTPPNAGR-GSAPRRG--SAALRYPRHFCSSLA 278
S G + TVP+ + + S P AG PP G GS P G SAA + +S
Sbjct: 3627 SPGVIKPPTATVPIQRHVPMPISAPEAGAPPTFGAIGSNPASGNNSAAAQAAAAAAASAM 3686
Query: 277 LGKPSPRGQSARAYAAKARGAGSLDLSQLQGSLQSPSNYSSSYTI 143
+ + Q+ + R G+ Q Q L P + +SS+ +
Sbjct: 3687 IDRQQQNLQNLQTLQNLQRMVGASQQQQPQQQLNYPMDPTSSFIV 3731
>gb|AAL65911.1|AF425651_1 multiple ankyrin repeat single KH domain protein [Drosophila
melanogaster]
Length = 4001
Score = 35.0 bits (79), Expect = 0.57
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = -1
Query: 448 STGSLDTAIETVPLLKLAAVQRSRPSAGTPPNAGR-GSAPRRG--SAALRYPRHFCSSLA 278
S G + TVP+ + + S P AG PP G GS P G SAA + +S
Sbjct: 3627 SPGVIKPPTATVPIQRHVPMPISAPEAGAPPTFGAIGSNPASGNNSAAAQAAAAAAASAM 3686
Query: 277 LGKPSPRGQSARAYAAKARGAGSLDLSQLQGSLQSPSNYSSSYTI 143
+ + Q+ + R G+ Q Q L P + +SS+ +
Sbjct: 3687 IDRQQQNLQNLQTLQNLQRMVGASQQQQPQQQLNYPMDPTSSFIV 3731
>gb|AAL68383.1| SD05950p [Drosophila melanogaster]
Length = 531
Score = 35.0 bits (79), Expect = 0.57
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = -1
Query: 448 STGSLDTAIETVPLLKLAAVQRSRPSAGTPPNAGR-GSAPRRG--SAALRYPRHFCSSLA 278
S G + TVP+ + + S P AG PP G GS P G SAA + +S
Sbjct: 157 SPGVIKPPTATVPIQRHVPMPISAPEAGAPPTFGAIGSNPASGNNSAAAQAAAAAAASAM 216
Query: 277 LGKPSPRGQSARAYAAKARGAGSLDLSQLQGSLQSPSNYSSSYTI 143
+ + Q+ + R G+ Q Q L P + +SS+ +
Sbjct: 217 IDRQQQNLQNLQTLQNLQRMVGASQQQQPQQQLNYPMDPTSSFIV 261
>gb|AAM11086.1| GH26759p [Drosophila melanogaster]
Length = 404
Score = 35.0 bits (79), Expect = 0.57
Identities = 29/105 (27%), Positives = 44/105 (41%), Gaps = 3/105 (2%)
Frame = -1
Query: 448 STGSLDTAIETVPLLKLAAVQRSRPSAGTPPNAGR-GSAPRRG--SAALRYPRHFCSSLA 278
S G + TVP+ + + S P AG PP G GS P G SAA + +S
Sbjct: 30 SPGVIKPPTATVPIQRHVPMPISAPEAGAPPTFGAIGSNPASGNNSAAAQAAAAAAASAM 89
Query: 277 LGKPSPRGQSARAYAAKARGAGSLDLSQLQGSLQSPSNYSSSYTI 143
+ + Q+ + R G+ Q Q L P + +SS+ +
Sbjct: 90 IDRQQQNLQNLQTLQNLQRMVGASQQQQPQQQLNYPMDPTSSFIV 134