Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000554A_C01 KCC000554A_c01
(518 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
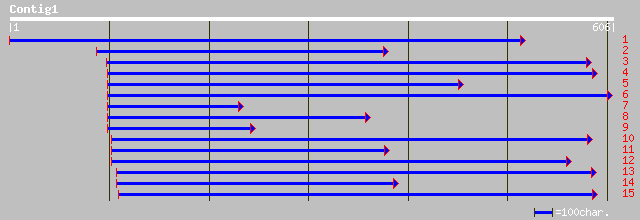
gb|AAF24819.1|AC007592_12 F12K11.22 [Arabidopsis thaliana] 111 5e-24
gb|AAL31897.1|AF419565_1 At1g06430/F12K11_24 [Arabidopsis thalia... 111 5e-24
gb|AAD17230.1| FtsH-like protein Pftf precursor [Nicotiana tabacum] 108 4e-23
ref|NP_850156.1| FtsH protease (VAR2) [Arabidopsis thaliana] gi|... 105 3e-22
emb|CAA09935.1| chloroplast protease [Capsicum annuum] 103 1e-21
>gb|AAF24819.1|AC007592_12 F12K11.22 [Arabidopsis thaliana]
Length = 662
Score = 111 bits (278), Expect = 5e-24
Identities = 62/105 (59%), Positives = 75/105 (71%), Gaps = 1/105 (0%)
Frame = +3
Query: 207 TRAVVVRAQQEQQTEVVASGKRDLIRNAVAAAVAMMPVMAAKA-EDAAGVASSRMSYSRF 383
++ VV+A +++ G L+ AA V ++ A A E GV+SSRMSYSRF
Sbjct: 31 SKVTVVKASLDEKKHEGRRGFFKLLLGNAAAGVGLLASGNANADEQGQGVSSSRMSYSRF 90
Query: 384 LEYLEMGRVKKVDLYENGTIAIVEAVSPELGNRVQRVRVQLPGTS 518
LEYL+ GRV+KVDLYENGTIAIVEAVSPELGNR+QRVRVQLPG S
Sbjct: 91 LEYLDKGRVEKVDLYENGTIAIVEAVSPELGNRIQRVRVQLPGLS 135
>gb|AAL31897.1|AF419565_1 At1g06430/F12K11_24 [Arabidopsis thaliana]
gi|27363292|gb|AAO11565.1| At1g06430/F12K11_24
[Arabidopsis thaliana] gi|28392858|gb|AAO41866.1|
putative FtsH protease [Arabidopsis thaliana]
Length = 685
Score = 111 bits (278), Expect = 5e-24
Identities = 62/105 (59%), Positives = 75/105 (71%), Gaps = 1/105 (0%)
Frame = +3
Query: 207 TRAVVVRAQQEQQTEVVASGKRDLIRNAVAAAVAMMPVMAAKA-EDAAGVASSRMSYSRF 383
++ VV+A +++ G L+ AA V ++ A A E GV+SSRMSYSRF
Sbjct: 31 SKVTVVKASLDEKKHEGRRGFFKLLLGNAAAGVGLLASGNANADEQGQGVSSSRMSYSRF 90
Query: 384 LEYLEMGRVKKVDLYENGTIAIVEAVSPELGNRVQRVRVQLPGTS 518
LEYL+ GRV+KVDLYENGTIAIVEAVSPELGNR+QRVRVQLPG S
Sbjct: 91 LEYLDKGRVEKVDLYENGTIAIVEAVSPELGNRIQRVRVQLPGLS 135
>gb|AAD17230.1| FtsH-like protein Pftf precursor [Nicotiana tabacum]
Length = 693
Score = 108 bits (270), Expect = 4e-23
Identities = 59/110 (53%), Positives = 74/110 (66%)
Frame = +3
Query: 189 LPGALRTRAVVVRAQQEQQTEVVASGKRDLIRNAVAAAVAMMPVMAAKAEDAAGVASSRM 368
LP + +T + V+A +Q+ + G L+ V V + D GV++SRM
Sbjct: 37 LPSSSKTSRIAVKASLQQRPDEGRRGFLKLLLGNVGLGVPALLGDGKAYADEQGVSNSRM 96
Query: 369 SYSRFLEYLEMGRVKKVDLYENGTIAIVEAVSPELGNRVQRVRVQLPGTS 518
SYSRFLEYL+ RV+KVDL+ENGTIAIVEA+SPELGNRVQRVRVQLPG S
Sbjct: 97 SYSRFLEYLDKDRVQKVDLFENGTIAIVEAISPELGNRVQRVRVQLPGLS 146
>ref|NP_850156.1| FtsH protease (VAR2) [Arabidopsis thaliana] gi|25308030|pir||F84714
probable ftsH chloroplast proteinase [imported] -
Arabidopsis thaliana gi|3201633|gb|AAC20729.1| FtsH
protease (VAR2) [Arabidopsis thaliana]
gi|7650138|gb|AAF65925.1|AF135189_1 zinc dependent
protease [Arabidopsis thaliana]
Length = 695
Score = 105 bits (262), Expect = 3e-22
Identities = 61/107 (57%), Positives = 74/107 (69%), Gaps = 1/107 (0%)
Frame = +3
Query: 201 LRTRAV-VVRAQQEQQTEVVASGKRDLIRNAVAAAVAMMPVMAAKAEDAAGVASSRMSYS 377
+RT V VV+A + + + G+RD ++ + A + D GV+SSRMSYS
Sbjct: 38 IRTSKVNVVKASLDGKKK--QEGRRDFLKILLGNAGVGLVASGKANADEQGVSSSRMSYS 95
Query: 378 RFLEYLEMGRVKKVDLYENGTIAIVEAVSPELGNRVQRVRVQLPGTS 518
RFLEYL+ RV KVDLYENGTIAIVEAVSPELGNRV+RVRVQLPG S
Sbjct: 96 RFLEYLDKDRVNKVDLYENGTIAIVEAVSPELGNRVERVRVQLPGLS 142
>emb|CAA09935.1| chloroplast protease [Capsicum annuum]
Length = 693
Score = 103 bits (257), Expect = 1e-21
Identities = 59/110 (53%), Positives = 72/110 (64%)
Frame = +3
Query: 189 LPGALRTRAVVVRAQQEQQTEVVASGKRDLIRNAVAAAVAMMPVMAAKAEDAAGVASSRM 368
LP + +T VVV+A +Q+ + G L+ V + D GV++SRM
Sbjct: 37 LPSSGKTSRVVVKASLQQRPDEGRRGFLKLLLGNVGLGAPALLGNGKAYADEQGVSNSRM 96
Query: 369 SYSRFLEYLEMGRVKKVDLYENGTIAIVEAVSPELGNRVQRVRVQLPGTS 518
SYS F EYL+ RV+KVDL+ENGTIAIVEAVSPELGNRVQRVRVQLPG S
Sbjct: 97 SYSIFSEYLDKDRVQKVDLFENGTIAIVEAVSPELGNRVQRVRVQLPGLS 146