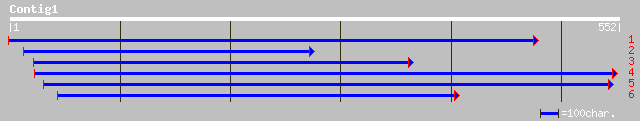
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000466A_C01 KCC000466A_c01
(552 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
gb|AAG10393.1| heavy-chain fibroin [Galleria mellonella] 44 0.001
gb|AAC47008.1| fibroin-1 gi|1589021|prf||2209442A fibroin:ISOTYPE=1 43 0.002
emb|CAA16808.1| EG:171E4.3 [Drosophila melanogaster] 43 0.003
ref|NP_569934.2| CG32809-PD [Drosophila melanogaster] gi|2283150... 43 0.003
gb|AAM52678.1| LD24507p [Drosophila melanogaster] 43 0.003
>gb|AAG10393.1| heavy-chain fibroin [Galleria mellonella]
Length = 1468
Score = 43.9 bits (102), Expect = 0.001
Identities = 44/121 (36%), Positives = 54/121 (44%), Gaps = 14/121 (11%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLA------- 355
+E SA SA AA S +S G+L PLG P + AG GAGL
Sbjct: 691 IEDGSSAASAAAAGSGASGVGGLGLGALGPLGGI--GPIGASSAGA-SGAGLGGVGAAGT 747
Query: 356 -----LGGNGASTTG--GAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTAAADS 514
+GG GAST G GAG + G G A AA+ AS DD S+AA+ +
Sbjct: 748 SGLGGIGGVGASTAGSAGAGLGGIGAGGSSGSSAASAASGASGAGEVIVIDDRSSAASAA 807
Query: 515 A 517
A
Sbjct: 808 A 808
Score = 43.5 bits (101), Expect = 0.002
Identities = 43/121 (35%), Positives = 52/121 (42%), Gaps = 14/121 (11%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLA------- 355
+E SA SA AA S +S G+L PLG + + T GAGL
Sbjct: 211 IEDGSSAASAAAAGSGASGVGGLGLGALGPLGGIGQSG---VSSATTSGAGLGGVGAVGA 267
Query: 356 -----LGGNGASTTG--GAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTAAADS 514
LGG GAS G GAG V G G A AA+ AS DD S+AA+ +
Sbjct: 268 SGLGGLGGAGASAAGSAGAGLGGVGVGGSSGSSAASAASGASGAGEVILIDDRSSAASRA 327
Query: 515 A 517
A
Sbjct: 328 A 328
Score = 39.7 bits (91), Expect = 0.027
Identities = 41/113 (36%), Positives = 48/113 (42%), Gaps = 11/113 (9%)
Frame = +2
Query: 212 SALSARAASSNSSSAQHDKAGSLRPLG--------CSFRQPRPLTPAGTFRGAGLA-LGG 364
SA SA AA S +S G L P G + L GT +GL LGG
Sbjct: 908 SAASAAAAGSGASGLGGLGLGGLGPYGGIGLNGVSSASALGAGLGGVGTAGASGLGGLGG 967
Query: 365 NGASTTG--GAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTAAADSA 517
GAS G GAG V G G A AA+ AS DD S+AA+ +A
Sbjct: 968 TGASAAGSAGAGLGGVGAGGSFGSSAASAASGASGAGEVIVIDDRSSAASAAA 1020
Score = 38.5 bits (88), Expect = 0.061
Identities = 41/113 (36%), Positives = 49/113 (43%), Gaps = 11/113 (9%)
Frame = +2
Query: 212 SALSARAASSNSSSAQHDKAGSLRPLG--------CSFRQPRPLTPAGTFRGAGL-ALGG 364
SA SA AASS +S G L P G + L GT +GL LGG
Sbjct: 428 SAASAAAASSGASGLGGLGLGGLGPYGGIGLNGVSSASALGAGLGGVGTAGASGLGGLGG 487
Query: 365 NGASTTG--GAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTAAADSA 517
G S G GAG V G G A AA+A S A +D S+AA+ +A
Sbjct: 488 AGVSAVGPAGAGLGGVGAGGSSGSSAASAASARSGPAPVIVIEDGSSAASAAA 540
Score = 35.8 bits (81), Expect = 0.40
Identities = 38/106 (35%), Positives = 42/106 (38%), Gaps = 11/106 (10%)
Frame = +2
Query: 212 SALSARAASSNSSSAQHDKAGSLRPLG--------CSFRQPRPLTPAGTFRGAGLA-LGG 364
SA SA AA S +S G L P G + L GT +GL LGG
Sbjct: 1363 SAASAAAAGSGASGLGGLGLGGLGPYGGIGLNGVSSASALGAGLGGVGTAGASGLGGLGG 1422
Query: 365 NGASTTG--GAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDS 496
GAS G GAG V G G A AA+ AS DD S
Sbjct: 1423 TGASAAGSAGAGLGGVGAGGSSGSSAASAASGASGAGEVIVIDDRS 1468
Score = 33.9 bits (76), Expect = 1.5
Identities = 33/113 (29%), Positives = 47/113 (41%), Gaps = 6/113 (5%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLALGGNGAS 376
+E SA SA AA S +S G+ PLG +G NG S
Sbjct: 1171 IEDGSSAASAAAAGSGASGLGGLGLGAWGPLG--------------------GIGPNGVS 1210
Query: 377 TTGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHP------DDDSTAAADSA 517
+ GS+A G+ G ++AASA + A P +D S+AA+ +A
Sbjct: 1211 SASATGSAAGSTGAGLGGSGAAGSSAASAASGAAGPAPVIVIEDGSSAASAAA 1263
Score = 33.9 bits (76), Expect = 1.5
Identities = 33/113 (29%), Positives = 47/113 (41%), Gaps = 6/113 (5%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLALGGNGAS 376
+E SA SA AA S +S G+ PLG +G NG S
Sbjct: 1090 IEDGSSAASAAAAGSGASGLGGLGLGAWGPLG--------------------GIGPNGVS 1129
Query: 377 TTGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHP------DDDSTAAADSA 517
+ GS+A G+ G ++AASA + A P +D S+AA+ +A
Sbjct: 1130 SASATGSAAGSTGAGLGGSGAAGSSAASAASGAAGPAPVIVIEDGSSAASAAA 1182
Score = 33.5 bits (75), Expect = 2.0
Identities = 31/98 (31%), Positives = 45/98 (45%), Gaps = 9/98 (9%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLG-CSFRQPRPLTPAGTFRGA-GLALGGNG 370
+E SA SA AA S +S G+ PLG + + G+ G+ G LGG+G
Sbjct: 529 IEDGSSAASAAAAGSGASGLGGLGLGAWGPLGGIGPNEVSSASATGSAAGSTGAGLGGSG 588
Query: 371 AS-------TTGGAGSSAVLHGSEGGWRAYGAAAAASA 463
A+ +G AG + V+ +G A AAA + A
Sbjct: 589 AAGSSAASAASGAAGPAPVIVIEDGSSAASAAAAGSGA 626
Score = 33.1 bits (74), Expect = 2.6
Identities = 37/113 (32%), Positives = 47/113 (40%), Gaps = 11/113 (9%)
Frame = +2
Query: 212 SALSARAASSNSSS------AQHDKAGSLRPLGCSFRQPRP--LTPAGTFRGAGLA-LGG 364
SA SA AA S +S G + P+G S L G +GL LGG
Sbjct: 802 SAASAAAAGSGASGPGGLGLGVWGPLGGIGPIGASSASASGAGLGGVGAAGTSGLGGLGG 861
Query: 365 NGASTTGGAGSSAVLHGSEG--GWRAYGAAAAASALARRAHPDDDSTAAADSA 517
GAS G AG+ G+ G G AA+ S DD S+AA+ +A
Sbjct: 862 AGASAAGSAGAGLGGIGASGSSGSSVASAASGTSGAGEVIVIDDRSSAASAAA 914
Score = 32.3 bits (72), Expect = 4.4
Identities = 38/121 (31%), Positives = 50/121 (40%), Gaps = 14/121 (11%)
Frame = +2
Query: 197 LEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLA------- 355
+E SA SA AA S +S +L PLG P ++ A GAGL
Sbjct: 1252 IEDGSSAASAAAAGSGASGVGGLGLSALGPLGGI--GPHGVSSASAL-GAGLGGVGAPGA 1308
Query: 356 --LGGNGASTTGGAGSS-----AVLHGSEGGWRAYGAAAAASALARRAHPDDDSTAAADS 514
LGG G + AGS+ V G G AA+ AS +D S+AA+ +
Sbjct: 1309 SGLGGLGVAGASAAGSAGAGLGGVGAGGSSGLSTASAASGASGAGEVIVINDRSSAASAA 1368
Query: 515 A 517
A
Sbjct: 1369 A 1369
Score = 31.6 bits (70), Expect = 7.5
Identities = 36/115 (31%), Positives = 49/115 (42%), Gaps = 13/115 (11%)
Frame = +2
Query: 212 SALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLA---------LGG 364
SA S AA S +S L P+G P T A T GAGL LGG
Sbjct: 322 SAASRAAAGSGASGVGGLGLSGLGPIGGI--GPIGATSAST-SGAGLGGVGAAGASGLGG 378
Query: 365 NGASTTGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHPD----DDSTAAADSA 517
G + AGS+ G G + G++AA++A + DD ++AA +A
Sbjct: 379 LGGAGASAAGSAGAGLGGIGAGGSSGSSAASAASGASGAGEVIVIDDRSSAASAA 433
>gb|AAC47008.1| fibroin-1 gi|1589021|prf||2209442A fibroin:ISOTYPE=1
Length = 360
Score = 43.1 bits (100), Expect = 0.002
Identities = 35/102 (34%), Positives = 43/102 (41%), Gaps = 11/102 (10%)
Frame = +2
Query: 191 GALEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLALG--- 361
G L A++ ASS +SSA S+R G PA F GAG G
Sbjct: 27 GRLLSNAGAITESTASSAASSASSTVTESIRTYG----------PAAIFSGAGAGAGVGV 76
Query: 362 --------GNGASTTGGAGSSAVLHGSEGGWRAYGAAAAASA 463
G GA GAG+ A G+ G + YGA AAA+A
Sbjct: 77 GGAGGYGQGYGAGAGAGAGAGAGAGGAGGYGQGYGAGAAAAA 118
Score = 32.0 bits (71), Expect = 5.7
Identities = 35/101 (34%), Positives = 38/101 (36%), Gaps = 1/101 (0%)
Frame = +2
Query: 179 GVSQGALEGALSALSARAASSNSSSAQHDKAGSLRPLGCSFRQPRPLTPAGTFRGAGLAL 358
G GA G S A Q AGS G + AG + G
Sbjct: 119 GAGAGAA-GGYGGGSGAGAGGAGGYGQGYGAGSGAGAGAAAAAGASAGAAGGYGGGAGVG 177
Query: 359 GGNGASTTGGAGSSAVLHGSEGGWRA-YGAAAAASALARRA 478
G GA GG G S +GS G A GAAAAA A AR A
Sbjct: 178 AGAGAGAAGGYGQS---YGSGAGAGAGAGAAAAAGAGARAA 215
Score = 32.0 bits (71), Expect = 5.7
Identities = 19/44 (43%), Positives = 22/44 (49%)
Frame = +2
Query: 338 RGAGLALGGNGASTTGGAGSSAVLHGSEGGWRAYGAAAAASALA 469
R AG GG GA GAG++A S G YG A A A+A
Sbjct: 213 RAAGGYGGGYGAGAGAGAGAAASAGASGGYGGGYGGGAGAGAVA 256
Score = 31.2 bits (69), Expect = 9.7
Identities = 22/51 (43%), Positives = 25/51 (48%)
Frame = +2
Query: 326 AGTFRGAGLALGGNGASTTGGAGSSAVLHGSEGGWRAYGAAAAASALARRA 478
AG GA GG + GGAG +G+ G A GAAAAA A A A
Sbjct: 118 AGAGAGAAGGYGGGSGAGAGGAGGYGQGYGAGSGAGA-GAAAAAGASAGAA 167
>emb|CAA16808.1| EG:171E4.3 [Drosophila melanogaster]
Length = 449
Score = 42.7 bits (99), Expect = 0.003
Identities = 35/101 (34%), Positives = 46/101 (44%), Gaps = 7/101 (6%)
Frame = +2
Query: 221 SARAASSNSSSAQHDKAGS--LRPLGCSFRQPRPLTPAG-----TFRGAGLALGGNGAST 379
S ASS+SSSAQ +GS LR LG + + + A A A+T
Sbjct: 323 SVDVASSSSSSAQDIASGSNTLRKLGSETNIQQKIAALSLACETSGAAAAAAAAAAAAAT 382
Query: 380 TGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTA 502
GG +S G+ GG GAA AA A A AH + +S +
Sbjct: 383 NGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNS 423
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/58 (32%), Positives = 26/58 (44%)
Frame = -3
Query: 499 SAVIVRVCAARQRRRGRGGAVRPPATFAAVKYGAGSSAAGGAGTVAAKRQASASKGSS 326
S AA GG V AT A+ G G+ AAG AG AA ++++ S+
Sbjct: 367 SGAAAAAAAAAAAAATNGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNSN 424
>ref|NP_569934.2| CG32809-PD [Drosophila melanogaster] gi|22831506|gb|AAF45655.2|
CG32809-PD [Drosophila melanogaster]
Length = 1234
Score = 42.7 bits (99), Expect = 0.003
Identities = 35/101 (34%), Positives = 46/101 (44%), Gaps = 7/101 (6%)
Frame = +2
Query: 221 SARAASSNSSSAQHDKAGS--LRPLGCSFRQPRPLTPAG-----TFRGAGLALGGNGAST 379
S ASS+SSSAQ +GS LR LG + + + A A A+T
Sbjct: 1108 SVDVASSSSSSAQDIASGSNTLRKLGSETNIQQKIAALSLACETSGAAAAAAAAAAAAAT 1167
Query: 380 TGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTA 502
GG +S G+ GG GAA AA A A AH + +S +
Sbjct: 1168 NGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNS 1208
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/58 (32%), Positives = 26/58 (44%)
Frame = -3
Query: 499 SAVIVRVCAARQRRRGRGGAVRPPATFAAVKYGAGSSAAGGAGTVAAKRQASASKGSS 326
S AA GG V AT A+ G G+ AAG AG AA ++++ S+
Sbjct: 1152 SGAAAAAAAAAAAAATNGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNSN 1209
>gb|AAM52678.1| LD24507p [Drosophila melanogaster]
Length = 653
Score = 42.7 bits (99), Expect = 0.003
Identities = 35/101 (34%), Positives = 46/101 (44%), Gaps = 7/101 (6%)
Frame = +2
Query: 221 SARAASSNSSSAQHDKAGS--LRPLGCSFRQPRPLTPAG-----TFRGAGLALGGNGAST 379
S ASS+SSSAQ +GS LR LG + + + A A A+T
Sbjct: 527 SVDVASSSSSSAQDIASGSNTLRKLGSETNIQQKIAALSLACETSGAAAAAAAAAAAAAT 586
Query: 380 TGGAGSSAVLHGSEGGWRAYGAAAAASALARRAHPDDDSTA 502
GG +S G+ GG GAA AA A A AH + +S +
Sbjct: 587 NGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNS 627
Score = 33.5 bits (75), Expect = 2.0
Identities = 19/58 (32%), Positives = 26/58 (44%)
Frame = -3
Query: 499 SAVIVRVCAARQRRRGRGGAVRPPATFAAVKYGAGSSAAGGAGTVAAKRQASASKGSS 326
S AA GG V AT A+ G G+ AAG AG AA ++++ S+
Sbjct: 571 SGAAAAAAAAAAAAATNGGVVASDATSGAIGGGGGAGAAGAAGASAAAAHSNSNSNSN 628