Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000461A_C01 KCC000461A_c01
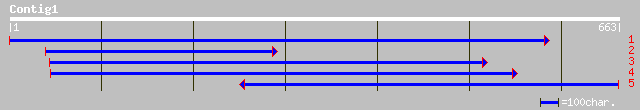
(538 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
sp|Q39586|METE_CHLRE 5-methyltetrahydropteroyltriglutamate--homo... 50 6e-14
gb|AAL40834.1|AF448220_1 BPLF1 [Human herpesvirus 4] 42 0.005
ref|NP_039850.1| Tegument protein [Human herpesvirus 4] gi|13557... 42 0.005
ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca] 42 0.007
ref|NP_767161.1| blr0521 [Bradyrhizobium japonicum] gi|27348769|... 35 0.013
>sp|Q39586|METE_CHLRE 5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (Methionine synthase, vitamin-B12
independent isozyme) (Cobalamin-independent methionine
synthase) gi|2129456|pir||S65083
5-methyltetrahydropteroyltriglutamate-homocysteine
S-methyltransferase (EC 2.1.1.14) - Chlamydomonas
reinhardtii gi|1017759|gb|AAC49178.1|
cobalamin-independent methionine synthase
gi|1587968|prf||2207381A Met synthase
Length = 814
Score = 49.7 bits (117), Expect(2) = 6e-14
Identities = 21/22 (95%), Positives = 21/22 (95%)
Frame = -3
Query: 467 GGAGSCRRNQPLRGLLPLSCLP 402
GGAG CRRNQPLRGLLPLSCLP
Sbjct: 793 GGAGQCRRNQPLRGLLPLSCLP 814
Score = 48.9 bits (115), Expect(2) = 6e-14
Identities = 24/26 (92%), Positives = 24/26 (92%)
Frame = -1
Query: 538 AELQLAGGAAVAPVAGGVEAAGKGAA 461
AELQLAGGAAVAPV GGVEAAGKG A
Sbjct: 770 AELQLAGGAAVAPVTGGVEAAGKGGA 795
>gb|AAL40834.1|AF448220_1 BPLF1 [Human herpesvirus 4]
Length = 3179
Score = 42.0 bits (97), Expect = 0.005
Identities = 45/158 (28%), Positives = 63/158 (39%), Gaps = 7/158 (4%)
Frame = +1
Query: 85 PIQTSVSRAALQGARMPPRLKLDIPSAHPNEPTCAPYTIQPEPALIRVTPGRA*PLSCTN 264
P+ T ++ + R+P ++ + +P A P+ P T P P T A + +
Sbjct: 514 PVST-IAPSVTPSPRLPLQIPIPLPQAAPSNPEIPLTTPSPSP-----TAAAAPTTTTLS 567
Query: 265 PCQPQPSYVPPFSSIPA*SVLLPSQYFCLVGTARTRSHDE*ISINQGRQLSGSSPRSGWF 444
P P PP S+ PA S LLP Q T + Q S + S
Sbjct: 568 P--PPTQQQPPQSAAPAPSPLLPQQQ----PTPSAAPAPSPLLPQQQPPPSAARAPSPLP 621
Query: 445 RRQLPAPPPCPQP---RRLPPQA----PLPPHPPAAAR 537
+Q P P P P ++LPP A P HPPAA R
Sbjct: 622 PQQQPLPSATPAPPPAQQLPPSATTLEPEKNHPPAADR 659
>ref|NP_039850.1| Tegument protein [Human herpesvirus 4] gi|135574|sp|P03186|TEGU_EBV
LARGE TEGUMENT PROTEIN gi|73929|pir||QQBE8 BPLF1 protein
- human herpesvirus 4 (strain B95-8)
gi|1334853|emb|CAA24839.1| BPLF1 reading frame, 1 NXT/S,
analogous to VZV RF22 [Human herpesvirus 4]
gi|23893598|emb|CAD53402.1| large tegument protein
[Human herpesvirus 4] gi|225109|prf||1208408E gene BPLF1
Length = 3149
Score = 42.0 bits (97), Expect = 0.005
Identities = 45/158 (28%), Positives = 63/158 (39%), Gaps = 7/158 (4%)
Frame = +1
Query: 85 PIQTSVSRAALQGARMPPRLKLDIPSAHPNEPTCAPYTIQPEPALIRVTPGRA*PLSCTN 264
P+ T ++ + R+P ++ + +P A P+ P T P P T A + +
Sbjct: 484 PVST-IAPSVTPSPRLPLQIPIPLPQAAPSNPKIPLTTPSPSP-----TAAAAPTTTTLS 537
Query: 265 PCQPQPSYVPPFSSIPA*SVLLPSQYFCLVGTARTRSHDE*ISINQGRQLSGSSPRSGWF 444
P P PP S+ PA S LLP Q T + Q S + S
Sbjct: 538 P--PPTQQQPPQSAAPAPSPLLPQQQ----PTPSAAPAPSPLLPQQQPPPSAARAPSPLP 591
Query: 445 RRQLPAPPPCPQP---RRLPPQA----PLPPHPPAAAR 537
+Q P P P P ++LPP A P HPPAA R
Sbjct: 592 PQQQPLPSATPAPPPAQQLPPSATTLEPEKNHPPAADR 629
>ref|ZP_00056820.1| COG2115: Xylose isomerase [Thermobifida fusca]
Length = 754
Score = 41.6 bits (96), Expect = 0.007
Identities = 37/141 (26%), Positives = 49/141 (34%), Gaps = 4/141 (2%)
Frame = +1
Query: 124 ARMPPRLKLDIPSAHPNEPTCAPYTIQPEPALIRVTPGRA*PLSCTNPCQPQPSYVPPFS 303
A PP P P PTC+P P+ P + P ++ P P+ P S
Sbjct: 61 AATPPAPATTTPPPTPTGPTCSP-----SPSAANSAPRESPPPPKSSAASPPPNSPPSTS 115
Query: 304 SIPA*SVLLPSQYFCLVGTARTRSHDE*ISINQGRQLSGSSPRSGWFRRQLPAPPPCPQP 483
S PA + P S + + SSP + P PP P P
Sbjct: 116 SAPAPATTWPPH-----------------SPSTPDPATSSSPSA-------PPEPPSPSP 151
Query: 484 RRLPPQAPLP----PHPPAAA 534
+ PP P P P PP A+
Sbjct: 152 KNPPPTPPAPSPDSPTPPDAS 172
Score = 31.2 bits (69), Expect = 9.0
Identities = 39/143 (27%), Positives = 46/143 (31%)
Frame = +1
Query: 97 SVSRAALQGARMPPRLKLDIPSAHPNEPTCAPYTIQPEPALIRVTPGRA*PLSCTNPCQP 276
S S A PP A + P+ P P P TP P S T P
Sbjct: 113 STSSAPAPATTWPPHSPSTPDPATSSSPSAPPEPPSPSPKNPPPTPPAPSPDSPTPPDAS 172
Query: 277 QPSYVPPFSSIPA*SVLLPSQYFCLVGTARTRSHDE*ISINQGRQLSGSSPRSGWFRRQL 456
PS P S+ PA S P + + S + S S P S
Sbjct: 173 SPSSAP--STQPASSPPEPLRS----ASPSKNSTNSPSPHPPAPAASSSCPTS-----TA 221
Query: 457 PAPPPCPQPRRLPPQAPLPPHPP 525
APP CP P +P P PP
Sbjct: 222 NAPPTCPTPPAASTASPEPTSPP 244
>ref|NP_767161.1| blr0521 [Bradyrhizobium japonicum] gi|27348769|dbj|BAC45786.1|
blr0521 [Bradyrhizobium japonicum USDA 110]
Length = 745
Score = 34.7 bits (78), Expect = 0.82
Identities = 16/26 (61%), Positives = 16/26 (61%), Gaps = 1/26 (3%)
Frame = +1
Query: 457 PAP-PPCPQPRRLPPQAPLPPHPPAA 531
PAP PP P P P AP PP PPAA
Sbjct: 146 PAPTPPAPPPAAAPQHAPPPPPPPAA 171
Score = 33.9 bits (76), Expect = 1.4
Identities = 16/29 (55%), Positives = 17/29 (58%), Gaps = 2/29 (6%)
Frame = +1
Query: 457 PAPPPCPQPRRLPPQ--APLPPHPPAAAR 537
P PP P P PP+ AP PP PP AAR
Sbjct: 77 PHPPAAPPPAAAPPRPAAPPPPPPPPAAR 105
Score = 33.9 bits (76), Expect = 1.4
Identities = 14/24 (58%), Positives = 14/24 (58%)
Frame = +1
Query: 460 APPPCPQPRRLPPQAPLPPHPPAA 531
APP P PP A PPHPPAA
Sbjct: 59 APPAAPARPAAPPPAAAPPHPPAA 82
Score = 32.7 bits (73), Expect = 3.1
Identities = 14/26 (53%), Positives = 15/26 (56%)
Frame = +1
Query: 460 APPPCPQPRRLPPQAPLPPHPPAAAR 537
APPP P P P P PP PPAA +
Sbjct: 94 APPPPPPPPAARPAPPPPPPPPAAPK 119
Score = 32.3 bits (72), Expect = 4.0
Identities = 37/151 (24%), Positives = 43/151 (27%)
Frame = +1
Query: 82 RPIQTSVSRAALQGARMPPRLKLDIPSAHPNEPTCAPYTIQPEPALIRVTPGRA*PLSCT 261
+P +T Q + PP P A P P P P P A P
Sbjct: 38 QPQETGPDGKPKQPPKGPPGAA---PPAAPARPAAPPPAAAPPHPPAAPPPAAAPPRPAA 94
Query: 262 NPCQPQPSYVPPFSSIPA*SVLLPSQYFCLVGTARTRSHDE*ISINQGRQLSGSSPRSGW 441
P P P P P P Q S P +
Sbjct: 95 PPPPPPPPAARPAPPPPPPPPAAPKQ--------------------------PSPPPAAA 128
Query: 442 FRRQLPAPPPCPQPRRLPPQAPLPPHPPAAA 534
++ P PPP P P AP PP PP AA
Sbjct: 129 PQQHAPTPPPPAPPAARP--APTPPAPPPAA 157
Score = 31.2 bits (69), Expect = 9.0
Identities = 14/26 (53%), Positives = 14/26 (53%)
Frame = +1
Query: 457 PAPPPCPQPRRLPPQAPLPPHPPAAA 534
PA P P P PP AP P PP AA
Sbjct: 102 PAARPAPPPPPPPPAAPKQPSPPPAA 127
Score = 31.2 bits (69), Expect = 9.0
Identities = 15/25 (60%), Positives = 16/25 (64%)
Frame = +3
Query: 447 AAAPSAAPLPAASTPPATGATAAPP 521
AAAPS P AA+TPP A APP
Sbjct: 370 AAAPSTVPGSAAATPPPNRAQFAPP 394
Score = 30.4 bits (67), Expect(2) = 0.013
Identities = 18/38 (47%), Positives = 19/38 (49%), Gaps = 7/38 (18%)
Frame = +3
Query: 426 STQRLVPAAAPSAAPL-------PAASTPPATGATAAP 518
ST PA P A P PAA +PPA GAT AP
Sbjct: 242 STPGAPPAGRPGAPPPGVRPGSPPAAGSPPAPGATPAP 279
Score = 29.3 bits (64), Expect(2) = 0.013
Identities = 18/55 (32%), Positives = 24/55 (42%), Gaps = 2/55 (3%)
Frame = +1
Query: 157 PSAHPNE-PTCAPYTIQPEPALIRVTPGRA*PLSCTNPC-QPQPSYVPPFSSIPA 315
P+A P PT P + P PA PG P + P P P+ P ++ PA
Sbjct: 185 PAARPTPAPTATPTPVAPPPAAPTARPGSPAPAATPAPTPTPAPTATPAPTATPA 239