Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
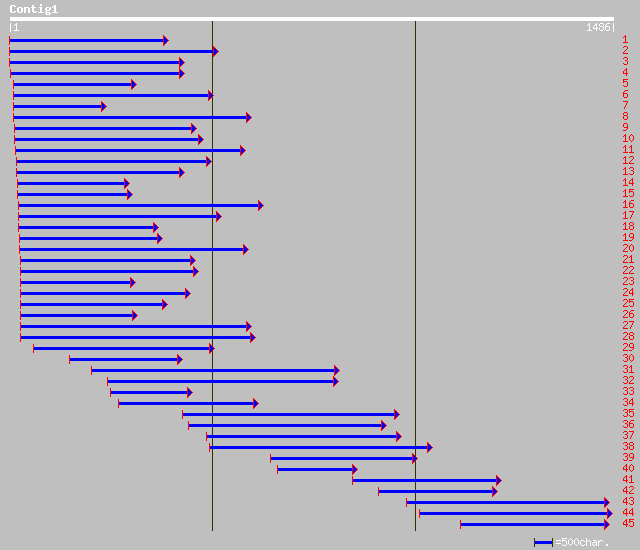
Query= KCC000459A_C01 KCC000459A_c01
(1468 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||S46234 Gbp1p protein - Chlamydomonas reinhardtii gi|520519|... 315 1e-84
gb|AAO39067.1| putative single stranded G-strand telomeric DNA-b... 94 4e-26
ref|XP_331180.1| hypothetical protein [Neurospora crassa] gi|289... 92 5e-25
emb|CAD60733.1| unnamed protein product [Podospora anserina] 87 2e-24
gb|AAM93968.1| Gbp1 [Griffithsia japonica] 92 2e-17
>pir||S46234 Gbp1p protein - Chlamydomonas reinhardtii gi|520519|gb|AAA21869.1|
Gbp1p
Length = 221
Score = 315 bits (808), Expect = 1e-84
Identities = 152/152 (100%), Positives = 152/152 (100%)
Frame = +1
Query: 55 MEDQSTIRLGKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFES 234
MEDQSTIRLGKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFES
Sbjct: 1 MEDQSTIRLGKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFES 60
Query: 235 PEEALHAIQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANG 414
PEEALHAIQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANG
Sbjct: 61 PEEALHAIQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANG 120
Query: 415 GRGAGRGGRGAGAPEATGESSGLQVVVQGIPW 510
GRGAGRGGRGAGAPEATGESSGLQVVVQGIPW
Sbjct: 121 GRGAGRGGRGAGAPEATGESSGLQVVVQGIPW 152
Score = 142 bits (359), Expect = 1e-32
Identities = 71/88 (80%), Positives = 75/88 (84%)
Frame = +2
Query: 455 PRRLASPAGCRLWSRASRGAYTWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTK 634
P +G ++ + AYTWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTK
Sbjct: 134 PEATGESSGLQVVVQGIPWAYTWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTK 193
Query: 635 EAAEAAVARYHESELEGRRLAVFIDRYQ 718
EAAEAAVARYHESELEGRRLAVFIDRYQ
Sbjct: 194 EAAEAAVARYHESELEGRRLAVFIDRYQ 221
Score = 60.1 bits (144), Expect = 1e-07
Identities = 31/78 (39%), Positives = 46/78 (58%)
Frame = +1
Query: 82 GKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQ 261
G + V + W +W++LKD F E G V +V+ DGRS+G+G V+F + E A A+
Sbjct: 142 GLQVVVQGIPWAYTWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVA 201
Query: 262 TLNGAELGGRRILVREDR 315
+ +EL GRR+ V DR
Sbjct: 202 RYHESELEGRRLAVFIDR 219
Score = 54.7 bits (130), Expect = 4e-06
Identities = 28/65 (43%), Positives = 41/65 (63%)
Frame = +2
Query: 518 TWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRRLA 697
+W++LKD F E G V +V+ DGRS+G+G V+F + E A A+ + +EL GRR+
Sbjct: 23 SWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQTLNGAELGGRRIL 82
Query: 698 VFIDR 712
V DR
Sbjct: 83 VREDR 87
Score = 37.7 bits (86), Expect = 0.53
Identities = 21/39 (53%), Positives = 21/39 (53%)
Frame = -3
Query: 461 ASGAPAPRPPRPAPRPPFAAPRPAPRAGRGARSAGAPGA 345
A GAPA R PRPA A AGRG R AGAP A
Sbjct: 98 APGAPAERAPRPARGAGRGAANGGRGAGRGGRGAGAPEA 136
>gb|AAO39067.1| putative single stranded G-strand telomeric DNA-binding protein
[Cryptosporidium parvum]
Length = 198
Score = 93.6 bits (231), Expect(2) = 4e-26
Identities = 57/142 (40%), Positives = 68/142 (47%), Gaps = 1/142 (0%)
Frame = +1
Query: 88 RCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQTL 267
R +VGNL WK W DLKD R+ GNV+ +V D+ GRS+G G+VE+ PEEA AI L
Sbjct: 8 RVYVGNLPWKAKWHDLKDHMRQAGNVIRADVFEDEVGRSRGCGVVEYSFPEEAQRAINEL 67
Query: 268 NGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANGGRGAG-RGGRG 444
N L R I VREDRED +R R RG G R
Sbjct: 68 NNTTLLDRLIFVREDREDES------------------SRYGRRSNKWNNRGYGMRTRTH 109
Query: 445 AGAPEATGESSGLQVVVQGIPW 510
A P E+ G QV V + W
Sbjct: 110 APRPPLKEENKGKQVFVTNLAW 131
Score = 67.8 bits (164), Expect = 5e-10
Identities = 35/78 (44%), Positives = 46/78 (58%)
Frame = +1
Query: 82 GKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQ 261
GK+ FV NLAWKT+ +DL F E G + V +DGRS+G + F P A A++
Sbjct: 121 GKQVFVTNLAWKTTQEDLAKAFNEIGPLESCEVFYFEDGRSRGIATIVFTDPNHAQLAVE 180
Query: 262 TLNGAELGGRRILVREDR 315
LN E+ GR ILVR D+
Sbjct: 181 KLNDREIDGREILVRIDQ 198
Score = 48.5 bits (114), Expect(2) = 4e-26
Identities = 24/65 (36%), Positives = 38/65 (57%)
Frame = +2
Query: 518 TWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRRLA 697
T +L F E+G ++ +V DGRSRG T+ FT A+ AV + ++ E++GR +
Sbjct: 134 TQEDLAKAFNEIGPLESCEVFYFEDGRSRGIATIVFTDPNHAQLAVEKLNDREIDGREIL 193
Query: 698 VFIDR 712
V ID+
Sbjct: 194 VRIDQ 198
Score = 47.8 bits (112), Expect = 5e-04
Identities = 28/83 (33%), Positives = 41/83 (48%)
Frame = +2
Query: 464 LASPAGCRLWSRASRGAYTWRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAA 643
+ S CR++ W +LKD + G V RADV GRSRG G V+++ E A
Sbjct: 1 MVSDKNCRVYVGNLPWKAKWHDLKDHMRQAGNVIRADVFEDEVGRSRGCGVVEYSFPEEA 60
Query: 644 EAAVARYHESELEGRRLAVFIDR 712
+ A+ + + L R + V DR
Sbjct: 61 QRAINELNNTTLLDRLIFVREDR 83
>ref|XP_331180.1| hypothetical protein [Neurospora crassa] gi|28920971|gb|EAA30296.1|
hypothetical protein [Neurospora crassa]
Length = 641
Score = 92.0 bits (227), Expect = 2e-17
Identities = 57/130 (43%), Positives = 70/130 (53%), Gaps = 7/130 (5%)
Frame = +1
Query: 82 GKRCFVGNLAWKTSWQDLKDKFREC---GNVVYTNVMRDDDGRSKGWGIVEFESPEEALH 252
G++ +V NL + WQDLKD FR+ G V+ +V DGR KG GIV FE+P++A +
Sbjct: 377 GRQIYVSNLPYNVGWQDLKDLFRQAARNGGVIRADVHIGPDGRPKGSGIVVFETPDDARN 436
Query: 253 AIQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGA----ANGGR 420
AIQ NG + GR + VREDR GA P R G GRGA GG
Sbjct: 437 AIQQFNGYDWQGRMLEVREDR-------FAGANMGPGGFGGRGGFGGGRGAFGGFGRGGF 489
Query: 421 GAGRGGRGAG 450
G GRGG G G
Sbjct: 490 GGGRGGFGGG 499
Score = 84.0 bits (206), Expect(2) = 5e-25
Identities = 56/156 (35%), Positives = 80/156 (50%), Gaps = 5/156 (3%)
Frame = +1
Query: 64 QSTIRLGKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEE 243
+ T + +R +VGNL++ W LKD R+ G V+Y +V+ +G SKG GIVE+ + E+
Sbjct: 248 RETSQQDRRVYVGNLSYDVKWHHLKDFMRQAGEVLYADVLLLPNGMSKGCGIVEYATREQ 307
Query: 244 ALHAIQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANGGRG 423
A +A+ TL+ L GR + DR+ + G PG A R G G G G G
Sbjct: 308 AQNAVATLSNQNLMGRLVY------DREAEPRFGPPGGGANRG-----GFGGGMGPNGYG 356
Query: 424 -----AGRGGRGAGAPEATGESSGLQVVVQGIPWGL 516
AG GG G G P A G G Q+ V +P+ +
Sbjct: 357 AVPGHAGPGGFG-GGPMAGG--GGRQIYVSNLPYNV 389
Score = 54.3 bits (129), Expect(2) = 5e-25
Identities = 29/68 (42%), Positives = 42/68 (61%), Gaps = 3/68 (4%)
Frame = +2
Query: 521 WRELKDMFAEV---GGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRR 691
W++LKD+F + GGV RADV G DGR +G G V F T + A A+ +++ + +GR
Sbjct: 391 WQDLKDLFRQAARNGGVIRADVHIGPDGRPKGSGIVVFETPDDARNAIQQFNGYDWQGRM 450
Query: 692 LAVFIDRY 715
L V DR+
Sbjct: 451 LEVREDRF 458
Score = 53.1 bits (126), Expect = 1e-05
Identities = 26/66 (39%), Positives = 37/66 (55%)
Frame = +1
Query: 94 FVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQTLNG 273
+V NL W TS +DL + F G V + + GRS+G G+V F++ + A AIQ G
Sbjct: 543 YVRNLPWSTSNEDLVELFSTIGKVEQAEIQYEPSGRSRGSGVVRFDNADTADTAIQKFQG 602
Query: 274 AELGGR 291
+ GGR
Sbjct: 603 YQYGGR 608
Score = 43.9 bits (102), Expect = 0.007
Identities = 23/56 (41%), Positives = 31/56 (55%)
Frame = +2
Query: 521 WRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGR 688
W LKD + G V ADV+ +G S+G G V++ T+E A+ AVA L GR
Sbjct: 268 WHHLKDFMRQAGEVLYADVLLLPNGMSKGCGIVEYATREQAQNAVATLSNQNLMGR 323
Score = 43.1 bits (100), Expect = 0.013
Identities = 19/63 (30%), Positives = 37/63 (58%)
Frame = +2
Query: 527 ELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRRLAVFI 706
+L ++F+ +G V++A++ GRSRG G V+F + A+ A+ ++ + GR L +
Sbjct: 555 DLVELFSTIGKVEQAEIQYEPSGRSRGSGVVRFDNADTADTAIQKFQGYQYGGRPLGLSF 614
Query: 707 DRY 715
+Y
Sbjct: 615 VKY 617
>emb|CAD60733.1| unnamed protein product [Podospora anserina]
Length = 516
Score = 86.7 bits (213), Expect = 1e-15
Identities = 60/137 (43%), Positives = 70/137 (50%), Gaps = 5/137 (3%)
Frame = +1
Query: 85 KRCFVGNLAWKTSWQDLKDKFREC---GNVVYTNVMRDDDGRSKGWGIVEFESPEEALHA 255
++ FV NL + WQDLKD FR+ G V+ +V DGR KG GIV FESP++A +A
Sbjct: 249 RQIFVSNLPFNIGWQDLKDLFRQSARTGGVIRADVHLGPDGRPKGSGIVVFESPDDARNA 308
Query: 256 IQTLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANGGRG--AG 429
IQ NG + GR I VREDR A G R RG G G GGRG G
Sbjct: 309 IQQFNGYDWQGRVIEVREDR----FANAGMGAGGYGGRGGFGGRG-GFGGGFGGRGGFGG 363
Query: 430 RGGRGAGAPEATGESSG 480
RGG G G G G
Sbjct: 364 RGGFGGGGYGRGGYGGG 380
Score = 84.0 bits (206), Expect(2) = 2e-24
Identities = 50/144 (34%), Positives = 72/144 (49%)
Frame = +1
Query: 85 KRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQT 264
+R +VGNL++ W LKD R+ G V+Y +V+ +G SKG GIVE+ + E+A +A+ T
Sbjct: 125 RRVYVGNLSYDVKWHHLKDFMRQAGEVLYADVLLLPNGMSKGCGIVEYATREQAQNAVAT 184
Query: 265 LNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANGGRGAGRGGRG 444
L+ L GR + DR+ + G PG A G G GA G G G G
Sbjct: 185 LSNQNLMGRLVY------DREAEPRFGPPGGGARGGFGGPVGPGGGAPYG--GGFNPGMG 236
Query: 445 AGAPEATGESSGLQVVVQGIPWGL 516
GAP Q+ V +P+ +
Sbjct: 237 GGAPGGGAPGGARQIFVSNLPFNI 260
Score = 52.4 bits (124), Expect(2) = 2e-24
Identities = 28/68 (41%), Positives = 42/68 (61%), Gaps = 3/68 (4%)
Frame = +2
Query: 521 WRELKDMF---AEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRR 691
W++LKD+F A GGV RADV G DGR +G G V F + + A A+ +++ + +GR
Sbjct: 262 WQDLKDLFRQSARTGGVIRADVHLGPDGRPKGSGIVVFESPDDARNAIQQFNGYDWQGRV 321
Query: 692 LAVFIDRY 715
+ V DR+
Sbjct: 322 IEVREDRF 329
Score = 50.8 bits (120), Expect = 6e-05
Identities = 25/66 (37%), Positives = 35/66 (52%)
Frame = +1
Query: 94 FVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQTLNG 273
+V NL W TS DL + F G V + + GRS+G G+V F++ + A AI G
Sbjct: 420 YVRNLPWSTSNDDLVELFSTIGKVEQAEIQYEPSGRSRGSGVVRFDNADTAETAINKFQG 479
Query: 274 AELGGR 291
+ GGR
Sbjct: 480 YQYGGR 485
Score = 43.9 bits (102), Expect = 0.007
Identities = 23/56 (41%), Positives = 31/56 (55%)
Frame = +2
Query: 521 WRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGR 688
W LKD + G V ADV+ +G S+G G V++ T+E A+ AVA L GR
Sbjct: 138 WHHLKDFMRQAGEVLYADVLLLPNGMSKGCGIVEYATREQAQNAVATLSNQNLMGR 193
Score = 43.5 bits (101), Expect = 0.010
Identities = 20/63 (31%), Positives = 37/63 (57%)
Frame = +2
Query: 527 ELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRRLAVFI 706
+L ++F+ +G V++A++ GRSRG G V+F + AE A+ ++ + GR L +
Sbjct: 432 DLVELFSTIGKVEQAEIQYEPSGRSRGSGVVRFDNADTAETAINKFQGYQYGGRPLGLSY 491
Query: 707 DRY 715
+Y
Sbjct: 492 VKY 494
>gb|AAM93968.1| Gbp1 [Griffithsia japonica]
Length = 156
Score = 91.7 bits (226), Expect(2) = 2e-17
Identities = 57/143 (39%), Positives = 70/143 (48%)
Frame = +1
Query: 82 GKRCFVGNLAWKTSWQDLKDKFRECGNVVYTNVMRDDDGRSKGWGIVEFESPEEALHAIQ 261
GKR +VGNL+W T WQ LKD RE G+VV+ V + GRS G GIVEFE+ E A AI
Sbjct: 24 GKRVYVGNLSWDTRWQGLKDHMREAGDVVHAEVFTEASGRSAGCGIVEFENSEGAETAIS 83
Query: 262 TLNGAELGGRRILVREDREDRDVKQADGAPGAPAERAPRPARGAGRGAANGGRGAGRGGR 441
TLN L GR I VR DRE+ G P+ GR G
Sbjct: 84 TLNDTMLDGRTIFVRADREE----------GKPSR--------------------GRYGD 113
Query: 442 GAGAPEATGESSGLQVVVQGIPW 510
G G+ G ++VV +P+
Sbjct: 114 GDGSFRGRPSDRGRKIVVWNLPY 136
Score = 52.8 bits (125), Expect(2) = 4e-06
Identities = 27/64 (42%), Positives = 37/64 (57%)
Frame = +2
Query: 521 WRELKDMFAEVGGVDRADVVTGYDGRSRGYGTVKFTTKEAAEAAVARYHESELEGRRLAV 700
W+ LKD E G V A+V T GRS G G V+F E AE A++ +++ L+GR + V
Sbjct: 38 WQGLKDHMREAGDVVHAEVFTEASGRSAGCGIVEFENSEGAETAISTLNDTMLDGRTIFV 97
Query: 701 FIDR 712
DR
Sbjct: 98 RADR 101
Score = 21.6 bits (44), Expect(2) = 4e-06
Identities = 9/31 (29%), Positives = 14/31 (45%)
Frame = +1
Query: 451 APEATGESSGLQVVVQGIPWGLHLARAEGHV 543
AP+ G S G +V V + W + H+
Sbjct: 15 APQTGGHSGGKRVYVGNLSWDTRWQGLKDHM 45
Score = 21.6 bits (44), Expect(2) = 2e-17
Identities = 7/15 (46%), Positives = 11/15 (72%)
Frame = +2
Query: 521 WRELKDMFAEVGGVD 565
W++LKD+F + G D
Sbjct: 140 WQDLKDLFRDYGERD 154