Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000336A_C01 KCC000336A_c01
(812 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
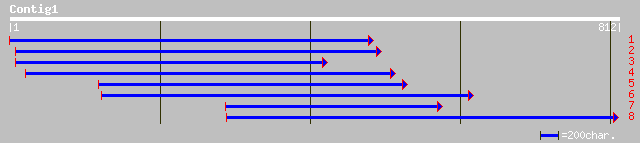
sp|Q39593|SAC1_CHLRE Putative sulfur deprivation response regula... 90 4e-17
ref|ZP_00013649.1| COG0645: Predicted kinase [Rhodospirillum rub... 47 4e-04
ref|ZP_00091011.1| COG2897: Rhodanese-related sulfurtransferase ... 45 0.001
gb|AAD31829.1| NapG oxidoreductase [Streptomyces collinus] 40 0.045
ref|ZP_00120241.1| COG0719: ABC-type transport system involved i... 40 0.045
>sp|Q39593|SAC1_CHLRE Putative sulfur deprivation response regulator
gi|2129463|pir||S69216 sulfur deprivation response
regulator sac1 - Chlamydomonas reinhardtii
gi|1197733|gb|AAB08008.1| sulfur deprivation response
regulator
Length = 585
Score = 90.1 bits (222), Expect = 4e-17
Identities = 47/47 (100%), Positives = 47/47 (100%)
Frame = +2
Query: 671 MIRGVVDADVCLFAASTLLLLRGIIQARDAFAGLANDSIVSIALMMM 811
MIRGVVDADVCLFAASTLLLLRGIIQARDAFAGLANDSIVSIALMMM
Sbjct: 1 MIRGVVDADVCLFAASTLLLLRGIIQARDAFAGLANDSIVSIALMMM 47
>ref|ZP_00013649.1| COG0645: Predicted kinase [Rhodospirillum rubrum]
Length = 759
Score = 47.0 bits (110), Expect = 4e-04
Identities = 45/123 (36%), Positives = 50/123 (40%), Gaps = 8/123 (6%)
Frame = +3
Query: 429 PLCRQTSASCSRGNCGTGKPSAG*K------SSAPRTSGRPSFRSAGPS-WSQGFSHRGS 587
P CR +SA + G PS+ SS P T GRPS S P W S R S
Sbjct: 11 PSCRPSSACHRTSSSSAGPPSSSPSCACSSPSSCPST-GRPSASSCRPCRWG---SFRRS 66
Query: 588 W-RWSSQRGAICPRTPGSRCGPPSRQWCP*SAAWWTPTCACSQPPRCCCCAASSKRATPS 764
W R SS R S G PS SAA WTP+ S P AASS + S
Sbjct: 67 WYRSSSASSGASSRPSSSASGRPSAS----SAASWTPSSGASWTPSSASFAASSAASGAS 122
Query: 765 RVW 773
W
Sbjct: 123 WRW 125
>ref|ZP_00091011.1| COG2897: Rhodanese-related sulfurtransferase [Azotobacter
vinelandii]
Length = 523
Score = 45.4 bits (106), Expect = 0.001
Identities = 52/179 (29%), Positives = 65/179 (36%), Gaps = 14/179 (7%)
Frame = +1
Query: 298 GAAELKHTQQRVAPCAERGCEQRSAEQH*YCKGKWRQARAPGAALYVARHP--RAVLEAT 471
G + + + R+ P A C+ H G+ R R P AA P R
Sbjct: 12 GGGQRRPDRHRLRPAARAPCQP-----HRPLPGRRRARRHPPAAAAEQGRPGERGKRRRP 66
Query: 472 VVPGNQAPAEKVPPQGRLGDP---HSGALDHPG------HRDFRIGALGAGAHSVERSVH 624
P PA ++PP G LG P H+G G R R G + AG S R H
Sbjct: 67 RRPARHLPASRLPPAGGLGPPGRRHAGTAGGAGRSCQRLRRPVRGGQVVAGQQSAARGRH 126
Query: 625 GRLVHAVDHLRGSGAHDPRRG---GRRRVPVRSLHAAAAARHHPSARRLRGSGQRLHCV 792
R +G H R RRR R+ A R P R+ RG RLH V
Sbjct: 127 PRRRPLGTDRQGHAHHHHRPAVPFSRRR---RTDRLARHPRVQPRPRQTRGCRGRLHRV 182
>gb|AAD31829.1| NapG oxidoreductase [Streptomyces collinus]
Length = 341
Score = 40.0 bits (92), Expect = 0.045
Identities = 34/117 (29%), Positives = 54/117 (46%), Gaps = 10/117 (8%)
Frame = +3
Query: 465 GNCGTGKPSAG*KSSAPRTSGRP---------SFRSAGPSWSQGFSHRGSWRWSSQRGAI 617
G+ G +P+A S++ TS P S R + PS + + R + RGA
Sbjct: 157 GSPGAARPAAAPWSTSAGTSWTPRPPCSARPRSTRPSAPSRTTSSTPASGPRLPAPRGAA 216
Query: 618 CPRTPGSRCGPPSRQ-WCP*SAAWWTPTCACSQPPRCCCCAASSKRATPSRVWPTTP 785
P +P + GP +R+ CP W+ + A PP C +AS+ +PS P++P
Sbjct: 217 SP-SPSAPVGPRTRRRTCP-----WSGSRAAPAPPNCAAPSASAPTGSPS---PSSP 264
>ref|ZP_00120241.1| COG0719: ABC-type transport system involved in Fe-S cluster
assembly, permease component [Bifidobacterium longum
DJO10A]
Length = 700
Score = 40.0 bits (92), Expect = 0.045
Identities = 37/108 (34%), Positives = 49/108 (45%), Gaps = 13/108 (12%)
Frame = +1
Query: 502 KVPPQGRL-GDPHSGAL--DHPGHRDFRIGALGAGAHSVERSVHGRLVHAVDHLRGSGAH 672
++ +GR G P +G L HPGH R A G + R++ GRL H D +RG A
Sbjct: 7 RIRAEGRACGHPAAGVLPHQHPGHGSVRAHAHHRGRRLL-RALRGRL-HRPDLVRGLAAR 64
Query: 673 DPRRGGRRRVPVRSLHAAA------AARHHPSARRLR----GSGQRLH 786
RR RR+ +LH A HP+ R R G G+R H
Sbjct: 65 RHRRDHRRKARPCALHHRAELVEQRLQPRHPACLRARGRHDGMGRRQH 112