Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000295A_C01 KCC000295A_c01
(639 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
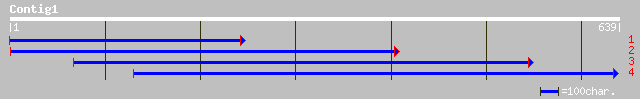
ref|NP_596410.1| DASS transporter of unknown specificity [Schizo... 76 4e-13
emb|CAD21226.1| related to PHO87 protein [Neurospora crassa] 74 1e-12
ref|XP_327979.1| hypothetical protein [Neurospora crassa] gi|289... 74 1e-12
gb|AAO32576.1| PHO87 [Saccharomyces kluyveri] 59 6e-08
emb|CAA96292.1| unnamed protein product [Saccharomyces cerevisiae] 58 1e-07
>ref|NP_596410.1| DASS transporter of unknown specificity [Schizosaccharomyces pombe]
gi|7492736|pir||T40336 probable MSF transporter -
fission yeast (Schizosaccharomyces pombe)
gi|2995339|emb|CAA18293.1| SPBC3B8.04c
[Schizosaccharomyces pombe]
Length = 867
Score = 76.3 bits (186), Expect = 4e-13
Identities = 43/132 (32%), Positives = 68/132 (50%), Gaps = 12/132 (9%)
Frame = +2
Query: 23 MKFTHQLKFNSVPEWREHYIQYGHLKKYIYALAKKEADLQAGGQDEEALLAPLLEAERDQ 202
MKF+H L+FN+VPEW E YI Y +LKK IY+L ++ LQ G DEE L E ++
Sbjct: 1 MKFSHSLQFNAVPEWSESYIAYSNLKKLIYSLEHEQITLQQGAPDEETRL-----LEHER 55
Query: 203 GPTEEGFQRELDAQLAATLSFFAVKEADLLAKVSALELDIQSLEK------------IPN 346
++ F LD +L + F+A KE ++ + ++ + ++ E P
Sbjct: 56 RSPDDRFMFALDKELQGIVEFYAPKEKEIADQYGRIKGEFETYENEYMSQGNNINYPTPE 115
Query: 347 RAEASTLARMGG 382
R + S+ +R G
Sbjct: 116 RLQKSSASRKSG 127
>emb|CAD21226.1| related to PHO87 protein [Neurospora crassa]
Length = 844
Score = 74.3 bits (181), Expect = 1e-12
Identities = 42/105 (40%), Positives = 61/105 (58%)
Frame = +2
Query: 23 MKFTHQLKFNSVPEWREHYIQYGHLKKYIYALAKKEADLQAGGQDEEALLAPLLEAERDQ 202
MKF+H ++FN+VP+W HYI Y +LKK IY L +K L +GG E PL++ E
Sbjct: 1 MKFSHSIQFNAVPDWSTHYIAYSNLKKLIYQL-EKVIHLSSGGDGES---RPLIQHE--- 53
Query: 203 GPTEEGFQRELDAQLAATLSFFAVKEADLLAKVSALELDIQSLEK 337
E F R LD +L LSF+ VKE +L +V + D+ + ++
Sbjct: 54 -DPEIVFVRALDVELEKVLSFYTVKERELFEEVQNVLRDVDAFDE 97
>ref|XP_327979.1| hypothetical protein [Neurospora crassa] gi|28917298|gb|EAA27007.1|
hypothetical protein [Neurospora crassa]
Length = 1006
Score = 74.3 bits (181), Expect = 1e-12
Identities = 42/105 (40%), Positives = 61/105 (58%)
Frame = +2
Query: 23 MKFTHQLKFNSVPEWREHYIQYGHLKKYIYALAKKEADLQAGGQDEEALLAPLLEAERDQ 202
MKF+H ++FN+VP+W HYI Y +LKK IY L +K L +GG E PL++ E
Sbjct: 161 MKFSHSIQFNAVPDWSTHYIAYSNLKKLIYQL-EKVIHLSSGGDGES---RPLIQHE--- 213
Query: 203 GPTEEGFQRELDAQLAATLSFFAVKEADLLAKVSALELDIQSLEK 337
E F R LD +L LSF+ VKE +L +V + D+ + ++
Sbjct: 214 -DPEIVFVRALDVELEKVLSFYTVKERELFEEVQNVLRDVDAFDE 257
>gb|AAO32576.1| PHO87 [Saccharomyces kluyveri]
Length = 889
Score = 58.9 bits (141), Expect = 6e-08
Identities = 33/81 (40%), Positives = 45/81 (54%), Gaps = 14/81 (17%)
Frame = +2
Query: 23 MKFTHQLKFNSVPEWREHYIQYGHLKKYIYALAKKEAD-LQAGGQDEEALLAPL------ 181
MKF+H LK+N+VPEW++HY+ Y LKK IY+L +E L GGQ + L L
Sbjct: 1 MKFSHSLKYNAVPEWQDHYLSYSQLKKLIYSLQARELQLLDDGGQLDRDRLKGLEGSSKA 60
Query: 182 -------LEAERDQGPTEEGF 223
++RD+G EE F
Sbjct: 61 LKKFKEKFMSKRDKGVGEEEF 81
>emb|CAA96292.1| unnamed protein product [Saccharomyces cerevisiae]
Length = 293
Score = 58.2 bits (139), Expect = 1e-07
Identities = 40/112 (35%), Positives = 57/112 (50%), Gaps = 8/112 (7%)
Frame = +2
Query: 23 MKFTHQLKFNSVPEWREHYIQYGHLKKYIYALAK-------KEADLQAGGQDEEALLAPL 181
MKF+H L+FNSVPEW Y+ Y LKK IY+L K K ++ ++E L PL
Sbjct: 1 MKFSHSLQFNSVPEWSTKYLAYSQLKKLIYSLQKDKLYSNNKHHVVEPHDANDENL--PL 58
Query: 182 L-EAERDQGPTEEGFQRELDAQLAATLSFFAVKEADLLAKVSALELDIQSLE 334
L +A D F L+ +L F+ +E L+A + L+ D+ LE
Sbjct: 59 LADASPDDQFYISKFVAALNQELKKIDKFYISQETGLIANYNELKDDVMELE 110