Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000201A_C03 KCC000201A_c03
(620 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
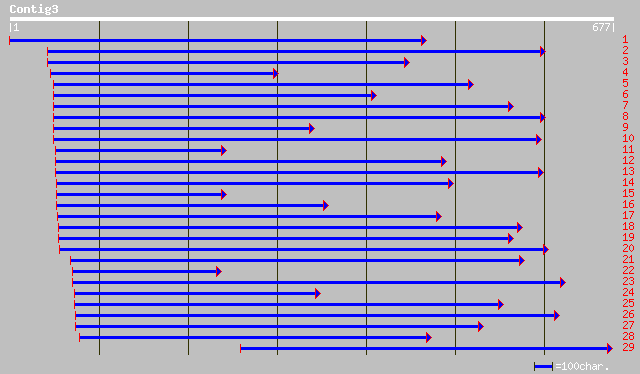
emb|CAA07265.1| alternative NADH-dehydrogenase [Yarrowia lipolyt... 63 4e-09
ref|NP_013586.1| NADH dehydrogenase (ubiquinone); Ndi1p [Sacchar... 61 1e-08
emb|CAB77710.2| NADH dehydrogenase [Candida albicans] 61 1e-08
ref|NP_180560.1| NADH dehydrogenase family [Arabidopsis thaliana... 60 2e-08
ref|XP_331372.1| hypothetical protein [Neurospora crassa] gi|289... 60 2e-08
>emb|CAA07265.1| alternative NADH-dehydrogenase [Yarrowia lipolytica]
Length = 582
Score = 62.8 bits (151), Expect = 4e-09
Identities = 47/136 (34%), Positives = 67/136 (48%), Gaps = 4/136 (2%)
Frame = +1
Query: 223 KAPATAPARTPLAPPSPAPAPGP-QYFPKPGDPQWPPQRPF---AAIALDNLLDSVVDVG 390
K TAPA P A G Q PKP D P ++ F ++ L ++ +G
Sbjct: 32 KIEGTAPAGLPKEVKQTAGHQGHHQEIPKP-DENHPRRKKFHFWRSLWRLTYLSAIASLG 90
Query: 391 RHLGRMNVEADKLAMRQQEQLVPGDNGRMRLRASTKPVVLVLGSGWGAHSLIKVIDTDTY 570
R+ V +R +P D +K ++VLGSGWG+ S +K +DT Y
Sbjct: 91 YIGYRIYV------IRNPSDQLPAD--------PSKKTLVVLGSGWGSVSFLKKLDTSNY 136
Query: 571 DVVVVSPRNHFLFTPM 618
+V+VVSPRN+FLFTP+
Sbjct: 137 NVIVVSPRNYFLFTPL 152
>ref|NP_013586.1| NADH dehydrogenase (ubiquinone); Ndi1p [Saccharomyces cerevisiae]
gi|417349|sp|P32340|NDI1_YEAST Rotenone-insensitive
NADH-ubiquinone oxidoreductase, mitochondrial precursor
(Internal NADH dehydrogenase) gi|283215|pir||S26704
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain NDI1
- yeast (Saccharomyces cerevisiae)
gi|805022|emb|CAA89160.1| Ndi1p [Saccharomyces
cerevisiae]
Length = 513
Score = 61.2 bits (147), Expect = 1e-08
Identities = 27/43 (62%), Positives = 35/43 (80%)
Frame = +1
Query: 490 STKPVVLVLGSGWGAHSLIKVIDTDTYDVVVVSPRNHFLFTPM 618
S KP VL+LGSGWGA S +K IDT Y+V ++SPR++FLFTP+
Sbjct: 51 SDKPNVLILGSGWGAISFLKHIDTKKYNVSIISPRSYFLFTPL 93
>emb|CAB77710.2| NADH dehydrogenase [Candida albicans]
Length = 574
Score = 61.2 bits (147), Expect = 1e-08
Identities = 26/41 (63%), Positives = 35/41 (84%)
Frame = +1
Query: 496 KPVVLVLGSGWGAHSLIKVIDTDTYDVVVVSPRNHFLFTPM 618
K +++LGSGWGA SL+K +DT Y+VV+VSPRN+FLFTP+
Sbjct: 99 KKTLVILGSGWGAISLLKNLDTTLYNVVIVSPRNYFLFTPL 139
>ref|NP_180560.1| NADH dehydrogenase family [Arabidopsis thaliana]
gi|7432659|pir||T02486 hypothetical protein At2g29990
[imported] - Arabidopsis thaliana
gi|3420052|gb|AAC31853.1| putative NADH dehydrogenase
(ubiquinone oxidoreductase) [Arabidopsis thaliana]
Length = 508
Score = 60.5 bits (145), Expect = 2e-08
Identities = 28/45 (62%), Positives = 33/45 (73%)
Frame = +1
Query: 484 RASTKPVVLVLGSGWGAHSLIKVIDTDTYDVVVVSPRNHFLFTPM 618
R KP V+VLGSGW L+K IDT+ YDVV VSPRNH +FTP+
Sbjct: 67 REGEKPRVVVLGSGWAGCRLMKGIDTNLYDVVCVSPRNHMVFTPL 111
>ref|XP_331372.1| hypothetical protein [Neurospora crassa] gi|28920404|gb|EAA29772.1|
hypothetical protein [Neurospora crassa]
Length = 577
Score = 60.5 bits (145), Expect = 2e-08
Identities = 24/42 (57%), Positives = 37/42 (87%)
Frame = +1
Query: 493 TKPVVLVLGSGWGAHSLIKVIDTDTYDVVVVSPRNHFLFTPM 618
+K ++VLG+GWG+ SL+K +DT+ Y+V+V+SPRN+FLFTP+
Sbjct: 111 SKKTLVVLGTGWGSVSLLKKLDTEHYNVIVISPRNYFLFTPL 152