Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
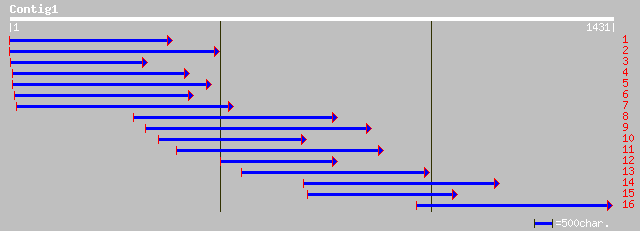
Query= KCC000189A_C01 KCC000189A_c01
(1431 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
emb|CAC84087.1| ZIK1 protein [Medicago sativa] 359 6e-98
dbj|BAB91130.1| Ser/Thr kinase [Arabidopsis thaliana] 337 2e-91
ref|NP_188505.1| protein kinase family [Arabidopsis thaliana] gi... 337 2e-91
dbj|BAB08432.1| MAP kinase [Arabidopsis thaliana] 337 2e-91
ref|NP_568599.1| protein kinase family [Arabidopsis thaliana] gi... 337 2e-91
>emb|CAC84087.1| ZIK1 protein [Medicago sativa]
Length = 591
Score = 359 bits (922), Expect = 6e-98
Identities = 168/266 (63%), Positives = 207/266 (77%)
Frame = +1
Query: 316 TTTYSRYDVVLGRGAFKTVFRAFDEEEGIEVAWNQIKVNDLASSPAERERLWAEIRVLKQ 495
T Y RY +LG+GAFK V+RAFDE EGIEVAWNQ+KV+DL + + ERL++E+ +LK
Sbjct: 22 TGRYGRYKEILGKGAFKKVYRAFDELEGIEVAWNQVKVSDLLRNSEDLERLYSEVHLLKT 81
Query: 496 LKHKNIMTFYDSWLDNKNNTVNFITELFTSGTLRQYRKKHKHIDEQVLKRWAWQILQGLV 675
LKHKNI+ FY+SW+D KN +NFITE+FTSGTLRQYRKKHKH+D + LK+W+ QIL+GL
Sbjct: 82 LKHKNIIKFYNSWVDTKNENINFITEIFTSGTLRQYRKKHKHVDLRALKKWSRQILEGLS 141
Query: 676 YLHGHNPPIIHRDLKCDNIFVNGTSGVIKIGDLGLVTLCRGFTAPQSVLGTPEFMAPELY 855
YLH HNPP+IHRDLKCDNIFVNG G +KIGDLGL + + T+ SV+GTPEFMAP LY
Sbjct: 142 YLHSHNPPVIHRDLKCDNIFVNGNQGEVKIGDLGLAAILQQATSAHSVIGTPEFMAPXLY 201
Query: 856 EEKYDEKVDVYSFGMCLLELATMEYPYSECKNAAQIYKKVTQGIHPGGLSKVEGQNLREF 1035
EE+Y+E VD+Y+FGMCLLEL T+EYPY EC NAAQIYKKVT GI P L+KV ++ F
Sbjct: 202 EEEYNELVDIYAFGMCLLELVTVEYPYVECANAAQIYKKVTSGIKPASLAKVNDPEVKAF 261
Query: 1036 IQVCIQHDPNQRPEARQLLKHPFFES 1113
I+ C H + P A+ LL PF +S
Sbjct: 262 IEKCTAHVTERLP-AKALLMDPFLQS 286
>dbj|BAB91130.1| Ser/Thr kinase [Arabidopsis thaliana]
Length = 550
Score = 337 bits (865), Expect = 2e-91
Identities = 159/260 (61%), Positives = 202/260 (77%)
Frame = +1
Query: 325 YSRYDVVLGRGAFKTVFRAFDEEEGIEVAWNQIKVNDLASSPAERERLWAEIRVLKQLKH 504
Y RYD VLGRGAFKTV++AFDE +GIEVAWN + + D+ P + ERL++E+ +LK LKH
Sbjct: 15 YIRYDDVLGRGAFKTVYKAFDEVDGIEVAWNLVSIEDVMQMPGQLERLYSEVHLLKALKH 74
Query: 505 KNIMTFYDSWLDNKNNTVNFITELFTSGTLRQYRKKHKHIDEQVLKRWAWQILQGLVYLH 684
+NI+ + SW+D KN T+N ITELFTSG+LR YRKKH+ +D + +K WA QIL+GL YLH
Sbjct: 75 ENIIKLFYSWVDEKNKTINMITELFTSGSLRVYRKKHRKVDPKAIKNWARQILKGLNYLH 134
Query: 685 GHNPPIIHRDLKCDNIFVNGTSGVIKIGDLGLVTLCRGFTAPQSVLGTPEFMAPELYEEK 864
NPP+IHRDLKCDNIFVNG +G +KIGDLGL T+ + TA +SV+GTPEFMAPELYEE+
Sbjct: 135 SQNPPVIHRDLKCDNIFVNGNTGEVKIGDLGLATVLQQPTA-RSVIGTPEFMAPELYEEE 193
Query: 865 YDEKVDVYSFGMCLLELATMEYPYSECKNAAQIYKKVTQGIHPGGLSKVEGQNLREFIQV 1044
Y+E VD+YSFGMC+LE+ T EYPY+EC+N AQIYKKVT I P L KV+ +R+FI+
Sbjct: 194 YNELVDIYSFGMCMLEMVTCEYPYNECRNQAQIYKKVTSNIKPQSLGKVDDPQVRQFIEK 253
Query: 1045 CIQHDPNQRPEARQLLKHPF 1104
C+ + RP A +L K PF
Sbjct: 254 CLL-PASSRPTALELSKDPF 272
>ref|NP_188505.1| protein kinase family [Arabidopsis thaliana]
gi|20302604|dbj|BAB91129.1| Ser/Thr kinase [Arabidopsis
thaliana]
Length = 567
Score = 337 bits (865), Expect = 2e-91
Identities = 157/265 (59%), Positives = 206/265 (77%)
Frame = +1
Query: 316 TTTYSRYDVVLGRGAFKTVFRAFDEEEGIEVAWNQIKVNDLASSPAERERLWAEIRVLKQ 495
T Y RY V+G+GAFKTV++AFDE +GIEVAWNQ++++D+ SP ERL++E+R+LK
Sbjct: 24 TFRYIRYKEVIGKGAFKTVYKAFDEVDGIEVAWNQVRIDDVLQSPNCLERLYSEVRLLKS 83
Query: 496 LKHKNIMTFYDSWLDNKNNTVNFITELFTSGTLRQYRKKHKHIDEQVLKRWAWQILQGLV 675
LKH NI+ FY+SW+D+KN TVN ITELFTSG+LR YRKKH+ ++ + +K WA QIL GL
Sbjct: 84 LKHNNIIRFYNSWIDDKNKTVNIITELFTSGSLRHYRKKHRKVNMKAVKNWARQILMGLR 143
Query: 676 YLHGHNPPIIHRDLKCDNIFVNGTSGVIKIGDLGLVTLCRGFTAPQSVLGTPEFMAPELY 855
YLHG PPIIHRDLKCDNIF+NG G +KIGDLGL T+ A +SV+GTPEFMAPELY
Sbjct: 144 YLHGQEPPIIHRDLKCDNIFINGNHGEVKIGDLGLATVMEQANA-KSVIGTPEFMAPELY 202
Query: 856 EEKYDEKVDVYSFGMCLLELATMEYPYSECKNAAQIYKKVTQGIHPGGLSKVEGQNLREF 1035
+E Y+E D+YSFGMC+LE+ T +YPY ECKN+AQIYKKV+ GI P LS+V+ +++F
Sbjct: 203 DENYNELADIYSFGMCMLEMVTFDYPYCECKNSAQIYKKVSSGIKPASLSRVKDPEVKQF 262
Query: 1036 IQVCIQHDPNQRPEARQLLKHPFFE 1110
I+ C+ ++R A++LL PF +
Sbjct: 263 IEKCLL-PASERLSAKELLLDPFLQ 286
>dbj|BAB08432.1| MAP kinase [Arabidopsis thaliana]
Length = 608
Score = 337 bits (865), Expect = 2e-91
Identities = 159/260 (61%), Positives = 202/260 (77%)
Frame = +1
Query: 325 YSRYDVVLGRGAFKTVFRAFDEEEGIEVAWNQIKVNDLASSPAERERLWAEIRVLKQLKH 504
Y RYD VLGRGAFKTV++AFDE +GIEVAWN + + D+ P + ERL++E+ +LK LKH
Sbjct: 73 YIRYDDVLGRGAFKTVYKAFDEVDGIEVAWNLVSIEDVMQMPGQLERLYSEVHLLKALKH 132
Query: 505 KNIMTFYDSWLDNKNNTVNFITELFTSGTLRQYRKKHKHIDEQVLKRWAWQILQGLVYLH 684
+NI+ + SW+D KN T+N ITELFTSG+LR YRKKH+ +D + +K WA QIL+GL YLH
Sbjct: 133 ENIIKLFYSWVDEKNKTINMITELFTSGSLRVYRKKHRKVDPKAIKNWARQILKGLNYLH 192
Query: 685 GHNPPIIHRDLKCDNIFVNGTSGVIKIGDLGLVTLCRGFTAPQSVLGTPEFMAPELYEEK 864
NPP+IHRDLKCDNIFVNG +G +KIGDLGL T+ + TA +SV+GTPEFMAPELYEE+
Sbjct: 193 SQNPPVIHRDLKCDNIFVNGNTGEVKIGDLGLATVLQQPTA-RSVIGTPEFMAPELYEEE 251
Query: 865 YDEKVDVYSFGMCLLELATMEYPYSECKNAAQIYKKVTQGIHPGGLSKVEGQNLREFIQV 1044
Y+E VD+YSFGMC+LE+ T EYPY+EC+N AQIYKKVT I P L KV+ +R+FI+
Sbjct: 252 YNELVDIYSFGMCMLEMVTCEYPYNECRNQAQIYKKVTSNIKPQSLGKVDDPQVRQFIEK 311
Query: 1045 CIQHDPNQRPEARQLLKHPF 1104
C+ + RP A +L K PF
Sbjct: 312 CLL-PASSRPTALELSKDPF 330
>ref|NP_568599.1| protein kinase family [Arabidopsis thaliana]
gi|15983509|gb|AAL11622.1|AF424629_1 AT5g41990/MJC20_9
[Arabidopsis thaliana] gi|21360469|gb|AAM47350.1|
AT5g41990/MJC20_9 [Arabidopsis thaliana]
Length = 563
Score = 337 bits (865), Expect = 2e-91
Identities = 159/260 (61%), Positives = 202/260 (77%)
Frame = +1
Query: 325 YSRYDVVLGRGAFKTVFRAFDEEEGIEVAWNQIKVNDLASSPAERERLWAEIRVLKQLKH 504
Y RYD VLGRGAFKTV++AFDE +GIEVAWN + + D+ P + ERL++E+ +LK LKH
Sbjct: 28 YIRYDDVLGRGAFKTVYKAFDEVDGIEVAWNLVSIEDVMQMPGQLERLYSEVHLLKALKH 87
Query: 505 KNIMTFYDSWLDNKNNTVNFITELFTSGTLRQYRKKHKHIDEQVLKRWAWQILQGLVYLH 684
+NI+ + SW+D KN T+N ITELFTSG+LR YRKKH+ +D + +K WA QIL+GL YLH
Sbjct: 88 ENIIKLFYSWVDEKNKTINMITELFTSGSLRVYRKKHRKVDPKAIKNWARQILKGLNYLH 147
Query: 685 GHNPPIIHRDLKCDNIFVNGTSGVIKIGDLGLVTLCRGFTAPQSVLGTPEFMAPELYEEK 864
NPP+IHRDLKCDNIFVNG +G +KIGDLGL T+ + TA +SV+GTPEFMAPELYEE+
Sbjct: 148 SQNPPVIHRDLKCDNIFVNGNTGEVKIGDLGLATVLQQPTA-RSVIGTPEFMAPELYEEE 206
Query: 865 YDEKVDVYSFGMCLLELATMEYPYSECKNAAQIYKKVTQGIHPGGLSKVEGQNLREFIQV 1044
Y+E VD+YSFGMC+LE+ T EYPY+EC+N AQIYKKVT I P L KV+ +R+FI+
Sbjct: 207 YNELVDIYSFGMCMLEMVTCEYPYNECRNQAQIYKKVTSNIKPQSLGKVDDPQVRQFIEK 266
Query: 1045 CIQHDPNQRPEARQLLKHPF 1104
C+ + RP A +L K PF
Sbjct: 267 CLL-PASSRPTALELSKDPF 285