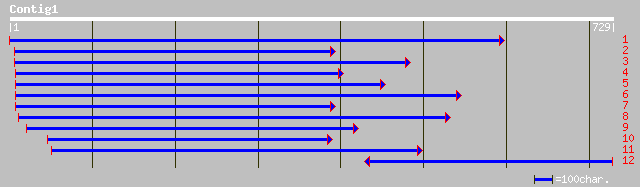
Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000071A_C01 KCC000071A_c01
(707 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
pir||T07905 low-carbon dioxide inducible protein 3 - Chlamydomon... 59 1e-07
dbj|BAB79394.1| ORF1 [TT virus] 43 0.005
gb|AAF82559.1|AF261761_2 putative capsid protein; pORF1 [TT virus] 41 0.007
ref|NP_808367.1| hypothetical protein LOC234686 [Mus musculus] g... 35 0.008
ref|XP_134529.3| similar to FH1/FH2 domains-containing protein (... 35 0.008
>pir||T07905 low-carbon dioxide inducible protein 3 - Chlamydomonas reinhardtii
gi|2367585|gb|AAB69691.1| low-carbon dioxide inducible
protein [Chlamydomonas reinhardtii]
Length = 118
Score = 58.5 bits (140), Expect = 1e-07
Identities = 43/87 (49%), Positives = 47/87 (53%), Gaps = 2/87 (2%)
Frame = +2
Query: 65 PWLPRTSRLRNPRSPPPCWASSPLPWSCSPPLPTPPRRPASARCWCSSNPTSKVCLKDSA 244
P RTSRLRNP +P CWASS P RR AS C PTSKVCLKDSA
Sbjct: 11 PVAARTSRLRNPVAPA-CWASSRCHGRVRPHCQR--RRDASRCCPRRPTPTSKVCLKDSA 67
Query: 245 K--N*IAWHGLDKTSRHWTSWQAVMAG 319
K N +AW G ++ QAVMAG
Sbjct: 68 KNSNCLAWLGQTSST---DKLQAVMAG 91
Score = 53.1 bits (126), Expect = 4e-06
Identities = 54/126 (42%), Positives = 66/126 (51%)
Frame = +1
Query: 34 MIAARVSSAKPVAAKNVQAAKPKVAPAVLGVFAAAMVVFAPIANAAETASVRQVLVLVQP 213
MIAARVSSAKPVAA+ + P VAPA V P AS R P
Sbjct: 1 MIAARVSSAKPVAARTSRLRNP-VAPACWASSRCHGRV-RPHCQRRRDAS-RCCPRRPTP 57
Query: 214 HLQGVPEGLRQELNCLAWLGQNFKALDKLASRDGGLSWTVT*LIPRGRARAERAVFQRWR 393
+ + + NCLAWLGQ + DKL + G +WTV ++ +GR AERAVFQ+
Sbjct: 58 TSKVCLKDSAKNSNCLAWLGQT-SSTDKLQAVMAG-TWTVD-MLSQGRT-AERAVFQK-G 112
Query: 394 ARVAGF 411
RVAGF
Sbjct: 113 PRVAGF 118
Score = 43.9 bits (102), Expect = 0.002
Identities = 20/28 (71%), Positives = 20/28 (71%)
Frame = +3
Query: 132 RCHGRVRPHCQRRRDGQRPPGAGARPTP 215
RCHGRVRPHCQRRRD R RPTP
Sbjct: 32 RCHGRVRPHCQRRRDASR--CCPRRPTP 57
>dbj|BAB79394.1| ORF1 [TT virus]
Length = 67
Score = 42.7 bits (99), Expect = 0.005
Identities = 28/63 (44%), Positives = 35/63 (55%), Gaps = 9/63 (14%)
Frame = -2
Query: 187 GRWPSRRRWQWGRTRPWQRRRRPARRG------RPWVSQPGRS---WQPRA*PRRREQRS 35
GRW RRRW R RPW+ RRR +RR R +VS+ GR W+ R RRR +R
Sbjct: 6 GRW-RRRRWGRRRRRPWRLRRRRSRRSFRRRGRRRYVSRRGRRRRFWRRRRRGRRRGRRK 64
Query: 34 FLR 26
L+
Sbjct: 65 RLK 67
>gb|AAF82559.1|AF261761_2 putative capsid protein; pORF1 [TT virus]
Length = 766
Score = 40.8 bits (94), Expect(2) = 0.007
Identities = 25/54 (46%), Positives = 29/54 (53%), Gaps = 5/54 (9%)
Frame = -2
Query: 184 RWPSRRRWQWGRTRPWQRRRRPAR-----RGRPWVSQPGRSWQPRA*PRRREQR 38
RWP RRR W R +RRRR AR R R W + GR W+ R RRR +R
Sbjct: 12 RWPRRRRTTWRRRPRPRRRRRTARTRRRGRVRRW-RRRGRGWRRRTYIRRRRRR 64
Score = 20.8 bits (42), Expect(2) = 0.007
Identities = 5/11 (45%), Positives = 5/11 (45%)
Frame = -1
Query: 221 WRWGWTSTSTW 189
WRW W W
Sbjct: 3 WRWWWRRRRRW 13
>ref|NP_808367.1| hypothetical protein LOC234686 [Mus musculus]
gi|26326725|dbj|BAC27106.1| unnamed protein product [Mus
musculus]
Length = 1197
Score = 34.7 bits (78), Expect(2) = 0.008
Identities = 21/50 (42%), Positives = 23/50 (46%)
Frame = +2
Query: 47 ASPRLSPWLPRTSRLRNPRSPPPCWASSPLPWSCSPPLPTPPRRPASARC 196
ASP LS P S P PPP P+ SC PP P P PA+ C
Sbjct: 572 ASPFLSSLSPSLSGGPPPPPPPP----PPITGSCPPPPPPPLPPPATGSC 617
Score = 26.6 bits (57), Expect(2) = 0.008
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 9/48 (18%)
Frame = +3
Query: 186 PPGAGARPTPPPRCA---------*RTPPRTKLLGMAWTKLQGTGQAG 302
PP G+ P PPP A R P + K + + W +L+ TG G
Sbjct: 626 PPIIGSCPPPPPLAAPFTHSALDGPRHPTKRKTVKLFWRELKLTGGPG 673
>ref|XP_134529.3| similar to FH1/FH2 domains-containing protein (Formin homolog
overexpressed in spleen) (FHOS) (Formin homology 2
domain containing 1) [Mus musculus]
Length = 1181
Score = 34.7 bits (78), Expect(2) = 0.008
Identities = 21/50 (42%), Positives = 23/50 (46%)
Frame = +2
Query: 47 ASPRLSPWLPRTSRLRNPRSPPPCWASSPLPWSCSPPLPTPPRRPASARC 196
ASP LS P S P PPP P+ SC PP P P PA+ C
Sbjct: 572 ASPFLSSLSPSLSGGPPPPPPPP----PPITGSCPPPPPPPLPPPATGSC 617
Score = 26.6 bits (57), Expect(2) = 0.008
Identities = 16/48 (33%), Positives = 22/48 (45%), Gaps = 9/48 (18%)
Frame = +3
Query: 186 PPGAGARPTPPPRCA---------*RTPPRTKLLGMAWTKLQGTGQAG 302
PP G+ P PPP A R P + K + + W +L+ TG G
Sbjct: 626 PPIIGSCPPPPPLAAPFTHSALDGPRHPTKRKTVKLFWRELKLTGGPG 673