Nr search
BLASTX 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= KCC000026A_C01 KCC000026A_c01
(988 letters)
Database: nr
1,537,769 sequences; 498,525,298 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
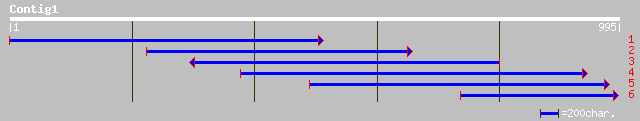
dbj|BAC87430.1| unnamed protein product [Homo sapiens] 44 0.003
dbj|BAC05810.1| seven transmembrane helix receptor [Homo sapiens] 44 0.004
ref|NP_042981.1| Hypothetical protein [Human herpesvirus 6] gi|8... 40 0.061
gb|AAL55824.1|AF318317_1 unknown [Homo sapiens] 40 0.061
dbj|BAC87611.1| unnamed protein product [Homo sapiens] 40 0.080
>dbj|BAC87430.1| unnamed protein product [Homo sapiens]
Length = 303
Score = 44.3 bits (103), Expect = 0.003
Identities = 27/103 (26%), Positives = 45/103 (43%), Gaps = 8/103 (7%)
Frame = -3
Query: 485 HTAVMTTTGQYAGCVGVPGVRTNHTLDHTHIHPALAPFPCVTLYLCAH-------CVKIH 327
HT + T T Y C+ +HT HTH++ + C+ +Y+C + CV +
Sbjct: 22 HTYIYTYTCMYV-CI------YSHTYIHTHVYVCVHIHICIHMYMCVYTHIYACTCVCVC 74
Query: 326 VEKETATCVCVCLCLSCKSLVAARIQCSLLE-ALGCVCLNVYV 201
V T TC+CVC+ + + + CVC + Y+
Sbjct: 75 VHTHTYTCICVCVHTHIYIHMCVCVYTRIYTYTCVCVCTHAYI 117
Score = 40.0 bits (92), Expect = 0.061
Identities = 25/104 (24%), Positives = 44/104 (42%), Gaps = 11/104 (10%)
Frame = -3
Query: 404 HTHIHPALAPFPCVTLYLCAHC-VKIHVEKETATCVCVCL----------CLSCKSLVAA 258
HTHI+ + C Y+ H V ++ T TC+CVC+ C+ + +
Sbjct: 190 HTHIYIHMYVCVCTHAYIHTHVYVCVYTRIYTYTCMCVCVYTRIYTYTYVCVCTHAYIHT 249
Query: 257 RIQCSLLEALGCVCLNVYVSRPFTITMNPFTFLLPTHCRQTINA 126
++ CVC + Y+ + + +T++ CR TI A
Sbjct: 250 QVYV-------CVCTHAYIHTHVCVCVYTYTYMYTLWCRGTIIA 286
>dbj|BAC05810.1| seven transmembrane helix receptor [Homo sapiens]
Length = 346
Score = 43.9 bits (102), Expect = 0.004
Identities = 23/62 (37%), Positives = 31/62 (49%), Gaps = 3/62 (4%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLS---CKSLVAARIQCSLLEALGCVCLNVYVS 198
C L LC C+++ V CVCVCLC+S C S C + CVC++V +S
Sbjct: 136 CACLSLCV-CIRVSVCLSLRVCVCVCLCVSVYVCVSSYLCVYLCVCMYVCECVCVSVSLS 194
Query: 197 RP 192
P
Sbjct: 195 DP 196
Score = 37.0 bits (84), Expect = 0.52
Identities = 21/67 (31%), Positives = 31/67 (45%), Gaps = 6/67 (8%)
Frame = -3
Query: 383 LAPFPCVTLYLCAH---CVKIHVEKETATCVCVCLCLS---CKSLVAARIQCSLLEALGC 222
L F CV +Y+ CV +H+ VC+C+C+S C S+ A C + C
Sbjct: 91 LCVFLCVCVYVSVSVCLCVYLHISVYLCVYVCICVCVSVSLCVSVCACLSLCVCIRVSVC 150
Query: 221 VCLNVYV 201
+ L V V
Sbjct: 151 LSLRVCV 157
Score = 36.2 bits (82), Expect = 0.89
Identities = 23/71 (32%), Positives = 33/71 (46%), Gaps = 14/71 (19%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCLS-------------CKSLVAARIQCSLLEA 231
C+ LY+C CV + V + CVC+C+C+S C S+ + C L
Sbjct: 16 CLYLYVCVCICVSVCVCICVSVCVCICVCVSVFVCVCICVCVCVCVSMCVSISVCVYLCL 75
Query: 230 LGCVCLNVYVS 198
CV + VYVS
Sbjct: 76 CVCVSVCVYVS 86
>ref|NP_042981.1| Hypothetical protein [Human herpesvirus 6]
gi|854065|emb|CAA58337.1| U88 [Human herpesvirus 6]
Length = 413
Score = 40.0 bits (92), Expect = 0.061
Identities = 21/57 (36%), Positives = 30/57 (51%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLSCKSLVAARIQCSLLEALGCVCLNVYVS 198
CV L +C CV + V CVCVC+CL C SL C + C+C+++ +S
Sbjct: 278 CVCLCVCL-CVCLCV------CVCVCVCLLCMSLCMCMCMCMCMCMCMCMCMSLCMS 327
Score = 35.0 bits (79), Expect = 2.0
Identities = 22/56 (39%), Positives = 26/56 (46%), Gaps = 2/56 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCLS-CKSLVAARIQCSLLEALGCVCLNV 207
CV L +C CV + V CVCVC+CL C L C + CVCL V
Sbjct: 228 CVCLCVCVCVCVCVCVCVCVCVCVCVCVCLCVCVCLCVCLCVCLCVCVCVCVCLCV 283
Score = 35.0 bits (79), Expect = 2.0
Identities = 17/55 (30%), Positives = 25/55 (44%), Gaps = 3/55 (5%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCL---SCKSLVAARIQCSLLEALGCVCL 213
CV +C C ++ CVCVC CL +C + A C+ L C+C+
Sbjct: 100 CVCARVCV-CARVCARVCVCACVCVCACLCVCACLCVCACLCVCACLCVCACLCV 153
Score = 34.7 bits (78), Expect = 2.6
Identities = 22/58 (37%), Positives = 28/58 (47%), Gaps = 2/58 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCL-SCKSLVAARIQCSLLEALGCVCLNVYV 201
C L +CA CV + CVC CLC+ +C + A C+ L CVCL V V
Sbjct: 124 CACLCVCACLCVCACLCVCACLCVCACLCVCACLCVCACLCVCACLCVCVCVCLCVCV 181
Score = 33.9 bits (76), Expect = 4.4
Identities = 22/58 (37%), Positives = 27/58 (45%), Gaps = 2/58 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCLS-CKSLVAARIQCSLLEALGCVCLNVYV 201
C L +CA CV + V C+CVC+CL C L C + CVCL V V
Sbjct: 160 CACLCVCACLCVCVCVCLCVCVCLCVCVCLCVCVCLCVCVCLCVCVCLCVCVCLCVCV 217
Score = 33.1 bits (74), Expect = 7.5
Identities = 20/57 (35%), Positives = 27/57 (47%), Gaps = 1/57 (1%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLS-CKSLVAARIQCSLLEALGCVCLNVYV 201
CV L +C C+ + V C+CVC+CL C L C + CVC+ V V
Sbjct: 192 CVCLCVCV-CLCVCVCLCVCVCLCVCVCLCVCVCLCVCVCLCVCVCVCVCVCVCVCV 247
Score = 32.7 bits (73), Expect = 9.8
Identities = 21/58 (36%), Positives = 27/58 (46%), Gaps = 2/58 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCLS-CKSLVAARIQCSLLEALGCVCLNVYV 201
CV L +C CV + + CVCVCLC+ C + C + CVCL V V
Sbjct: 204 CVCLCVCVCLCVCVCLCVCVCLCVCVCLCVCVCVCVCVCVCVCVCVCVCVCVCLCVCV 261
>gb|AAL55824.1|AF318317_1 unknown [Homo sapiens]
Length = 400
Score = 40.0 bits (92), Expect = 0.061
Identities = 18/59 (30%), Positives = 31/59 (52%), Gaps = 3/59 (5%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLSCKSLVAARIQ---CSLLEALGCVCLNVYV 201
C+ +Y+C CV +HV VC+C+C+ V A + C + C+C+++YV
Sbjct: 27 CMCVYVCV-CVHVHVCASVCAFVCMCVCVCVCGSVCAFVSVHVCMRVYVCVCICVHLYV 84
Score = 35.4 bits (80), Expect = 1.5
Identities = 18/64 (28%), Positives = 33/64 (51%), Gaps = 7/64 (10%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLSCKSLVAARI-------QCSLLEALGCVCLN 210
C+ +Y+C C+ +H+ CVC+C C+ + R+ C+ + A C+C+
Sbjct: 69 CMRVYVCV-CICVHLY--VCVCVCLCACVYACVCMCLRLCVRLCVSVCACVYACICMCIC 125
Query: 209 VYVS 198
V+VS
Sbjct: 126 VHVS 129
Score = 32.7 bits (73), Expect = 9.8
Identities = 15/58 (25%), Positives = 30/58 (50%), Gaps = 2/58 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCLSCKSLVAARIQ-CSLLEALGCVCLNVYV 201
CV + +C H C+++ + C+CV +C+ + V A + C + CV + V++
Sbjct: 141 CVCVCVCVHVCMRVCAHQCLHVCICVYVCVCARICVCACVYVCVCISVCACVYVCVHL 198
Score = 32.7 bits (73), Expect = 9.8
Identities = 16/54 (29%), Positives = 24/54 (43%), Gaps = 3/54 (5%)
Frame = -3
Query: 368 CVTLYLCAH---CVKIHVEKETATCVCVCLCLSCKSLVAARIQCSLLEALGCVC 216
C +Y+C H C +H CVCVC C+ A + +L+ C+C
Sbjct: 189 CACVYVCVHLCVCACVHA------CVCVCACVHLCVCACAHVGATLV----CIC 232
>dbj|BAC87611.1| unnamed protein product [Homo sapiens]
Length = 201
Score = 39.7 bits (91), Expect = 0.080
Identities = 19/52 (36%), Positives = 25/52 (47%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLSCKSLVAARIQCSLLEALGCVCL 213
CV + +C CV + V CVCVC+CL L + C + CVCL
Sbjct: 13 CVCVCVCVLCVSVCVCVCLCVCVCVCVCLCLCVLSVSVCLCVCVSVCLCVCL 64
Score = 34.3 bits (77), Expect = 3.4
Identities = 19/56 (33%), Positives = 26/56 (45%)
Frame = -3
Query: 368 CVTLYLCAHCVKIHVEKETATCVCVCLCLSCKSLVAARIQCSLLEALGCVCLNVYV 201
CV L LC V + + CV VCLC+ S+ C + CVCL++ V
Sbjct: 38 CVCLCLCVLSVSVCL----CVCVSVCLCVCLVSVCLCVCLCVCVSVCLCVCLHLCV 89
Score = 33.5 bits (75), Expect = 5.8
Identities = 14/37 (37%), Positives = 19/37 (50%)
Frame = -3
Query: 317 ETATCVCVCLCLSCKSLVAARIQCSLLEALGCVCLNV 207
E CVCVC+C+ C S+ C + C+CL V
Sbjct: 9 EGCVCVCVCVCVLCVSVCVCVCLCVCVCVCVCLCLCV 45
Score = 32.7 bits (73), Expect = 9.8
Identities = 14/30 (46%), Positives = 18/30 (59%), Gaps = 1/30 (3%)
Frame = -3
Query: 368 CVTLYLCAH-CVKIHVEKETATCVCVCLCL 282
CV L LC CV + V + CVCVC+C+
Sbjct: 158 CVCLCLCVFLCVCVFVSVCVSVCVCVCMCI 187