

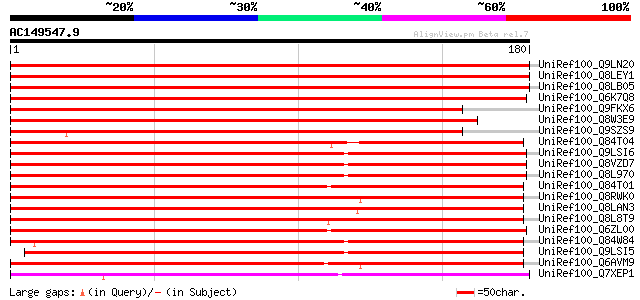
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149547.9 + phase: 0
(180 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LN20 F14O10.12 protein [Arabidopsis thaliana] 291 4e-78
UniRef100_Q8LEY1 Putative prolyl 4-hydroxylase, alpha subunit [A... 290 1e-77
UniRef100_Q8LB05 Putative prolyl 4-hydroxylase, alpha subunit [A... 278 6e-74
UniRef100_Q6K7Q8 Putative prolyl 4-hydroxylase [Oryza sativa] 275 3e-73
UniRef100_Q9FKX6 Prolyl 4-hydroxylase, alpha subunit-like protei... 265 4e-70
UniRef100_Q8W3E9 Putative prolyl 4-hydroxylase, alpha subunit [O... 234 6e-61
UniRef100_Q9SZS9 Hypothetical protein AT4g35810 [Arabidopsis tha... 220 1e-56
UniRef100_Q84T04 Putative oxidoreductase [Oryza sativa] 199 4e-50
UniRef100_Q9LSI6 Prolyl 4-hydroxylase alpha subunit-like protein... 196 3e-49
UniRef100_Q8VZD7 AT3g28480/MFJ20_16 [Arabidopsis thaliana] 196 3e-49
UniRef100_Q8L970 Prolyl 4-hydroxylase, putative [Arabidopsis tha... 196 3e-49
UniRef100_Q84T01 Putative oxidoreductase [Oryza sativa] 191 6e-48
UniRef100_Q8RWK0 Hypothetical protein At5g18900 [Arabidopsis tha... 190 2e-47
UniRef100_Q8LAN3 Prolyl 4-hydroxylase alpha subunit-like protein... 189 4e-47
UniRef100_Q8L8T9 Prolyl 4-hydroxylase alpha subunit-like protein... 187 8e-47
UniRef100_Q6ZL00 Prolyl 4-hydroxylase alpha-1 subunit-like prote... 187 1e-46
UniRef100_Q84W84 Putative prolyl 4-hydroxylase [Arabidopsis thal... 186 3e-46
UniRef100_Q9LSI5 Gb|AAF08583.1 [Arabidopsis thaliana] 185 5e-46
UniRef100_Q6AVM9 Putative prolyl 4-hydroxylase alpha subunit [Or... 184 1e-45
UniRef100_Q7XEP1 Hypothetical protein [Oryza sativa] 179 4e-44
>UniRef100_Q9LN20 F14O10.12 protein [Arabidopsis thaliana]
Length = 287
Score = 291 bits (746), Expect = 4e-78
Identities = 132/180 (73%), Positives = 153/180 (84%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+D ETG DSR RTSSG FL+RG D+I+K IE+RIAD+TFIP +HGE VLH
Sbjct: 108 MVKSTVVDSETGKSKDSRVRTSSGTFLRRGRDKIIKTIEKRIADYTFIPADHGEGLQVLH 167
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPHYDYF+D F+T GQR+ATMLMYLSDVEEGGETVFP A NFSSVPW+NE
Sbjct: 168 YEAGQKYEPHYDYFVDEFNTKNGGQRMATMLMYLSDVEEGGETVFPAANMNFSSVPWYNE 227
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFKI 180
LS+CGK GLS+KP+MG+A+LFWSM+PDATLDP+SLHG CPVI+G+KW KWMHVGE+KI
Sbjct: 228 LSECGKKGLSVKPRMGDALLFWSMRPDATLDPTSLHGGCPVIRGNKWSSTKWMHVGEYKI 287
>UniRef100_Q8LEY1 Putative prolyl 4-hydroxylase, alpha subunit [Arabidopsis thaliana]
Length = 287
Score = 290 bits (742), Expect = 1e-77
Identities = 131/180 (72%), Positives = 153/180 (84%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+D ETG DSR RTSSG FL+RG D+I+K IE+RIAD+TFIP +HGE VLH
Sbjct: 108 MVKSTVVDSETGKSKDSRVRTSSGTFLRRGRDKIIKTIEKRIADYTFIPADHGEGLQVLH 167
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPHYDYF+D F+T GQR+ATMLMYLSDVEEGGETVFP A NFSSVPW+NE
Sbjct: 168 YEAGQKYEPHYDYFVDEFNTKNGGQRMATMLMYLSDVEEGGETVFPAANMNFSSVPWYNE 227
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFKI 180
LS+CGK GLS+KP+MG+A+LFWSM+PDATLDP+SLHG CPVI+G+KW KW+HVGE+KI
Sbjct: 228 LSECGKKGLSVKPRMGDALLFWSMRPDATLDPTSLHGGCPVIRGNKWSSTKWIHVGEYKI 287
>UniRef100_Q8LB05 Putative prolyl 4-hydroxylase, alpha subunit [Arabidopsis thaliana]
Length = 291
Score = 278 bits (710), Expect = 6e-74
Identities = 125/180 (69%), Positives = 150/180 (82%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+DE+TG DSR RTSSG FL+RG D +V+ IE+RI+DFTFIPVE+GE VLH
Sbjct: 112 MVKSTVVDEKTGGSKDSRVRTSSGTFLRRGHDEVVEVIEKRISDFTFIPVENGEGLQVLH 171
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
Y+VGQKYEPHYDYF+D F+T GQRIAT+LMYLSDV++GGETVFP A+GN S+VPWWNE
Sbjct: 172 YQVGQKYEPHYDYFLDEFNTKNGGQRIATVLMYLSDVDDGGETVFPAARGNISAVPWWNE 231
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFKI 180
LS CGK GLS+ PK +A+LFW+M+PDA+LDPSSLHG CPV+KG+KW KW HV EFK+
Sbjct: 232 LSKCGKEGLSVLPKXRDALLFWNMRPDASLDPSSLHGGCPVVKGNKWSSTKWFHVHEFKV 291
>UniRef100_Q6K7Q8 Putative prolyl 4-hydroxylase [Oryza sativa]
Length = 310
Score = 275 bits (704), Expect = 3e-73
Identities = 123/179 (68%), Positives = 150/179 (83%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+D TG DSR RTSSG FL+RG D++++ IE+RIAD+TFIP+EHGE VLH
Sbjct: 131 MVKSTVVDSTTGKSKDSRVRTSSGMFLQRGRDKVIRAIEKRIADYTFIPMEHGEGLQVLH 190
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YEVGQKYEPH+DYF+D ++T GQR+AT+LMYLSDVEEGGET+FP+A N SS+PW+NE
Sbjct: 191 YEVGQKYEPHFDYFLDEYNTKNGGQRMATLLMYLSDVEEGGETIFPDANVNSSSLPWYNE 250
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
LS+C + GL++KPKMG+A+LFWSMKPDATLDP SLHG CPVIKG+KW KWMHV E+K
Sbjct: 251 LSECARKGLAVKPKMGDALLFWSMKPDATLDPLSLHGGCPVIKGNKWSSTKWMHVREYK 309
>UniRef100_Q9FKX6 Prolyl 4-hydroxylase, alpha subunit-like protein [Arabidopsis
thaliana]
Length = 267
Score = 265 bits (677), Expect = 4e-70
Identities = 120/157 (76%), Positives = 137/157 (86%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+DE+TG DSR RTSSG FL RG D+ ++ IE+RI+DFTFIPVEHGE VLH
Sbjct: 110 MEKSTVVDEKTGKSTDSRVRTSSGTFLARGRDKTIREIEKRISDFTFIPVEHGEGLQVLH 169
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE+GQKYEPHYDYFMD ++T GQRIAT+LMYLSDVEEGGETVFP AKGN+S+VPWWNE
Sbjct: 170 YEIGQKYEPHYDYFMDEYNTRNGGQRIATVLMYLSDVEEGGETVFPAAKGNYSAVPWWNE 229
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHG 157
LS+CGKGGLS+KPKMG+A+LFWSM PDATLDPSSLHG
Sbjct: 230 LSECGKGGLSVKPKMGDALLFWSMTPDATLDPSSLHG 266
>UniRef100_Q8W3E9 Putative prolyl 4-hydroxylase, alpha subunit [Oryza sativa]
Length = 343
Score = 234 bits (598), Expect = 6e-61
Identities = 107/162 (66%), Positives = 133/162 (82%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V+D TG DSR RTSSG FL RG D+I++ IE+RI+D+TFIPVE+GE VLH
Sbjct: 142 MKKSTVVDASTGGSKDSRVRTSSGMFLGRGQDKIIRTIEKRISDYTFIPVENGEGLQVLH 201
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YEVGQKYEPH+DYF D F+T GQRIAT+LMYLSDVEEGGET+FP++K N SS P++NE
Sbjct: 202 YEVGQKYEPHFDYFHDEFNTKNGGQRIATLLMYLSDVEEGGETIFPSSKANSSSSPFYNE 261
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVI 162
LS+C K GL++KPKMG+A+LFWSM+PD +LD +SLHG P++
Sbjct: 262 LSECAKKGLAVKPKMGDALLFWSMRPDGSLDATSLHGEIPIL 303
>UniRef100_Q9SZS9 Hypothetical protein AT4g35810 [Arabidopsis thaliana]
Length = 307
Score = 220 bits (561), Expect = 1e-56
Identities = 111/188 (59%), Positives = 127/188 (67%), Gaps = 31/188 (16%)
Query: 1 MHKSAVIDEETGNGVDSR-------------------------------ERTSSGAFLKR 29
M KS V+D +TG +DSR RTSSG FL R
Sbjct: 112 MMKSKVVDVKTGKSIDSRFCTLTSVVVFTFQLNLERFENSKFANPSLCRVRTSSGTFLNR 171
Query: 30 GSDRIVKNIERRIADFTFIPVEHGENFNVLHYEVGQKYEPHYDYFMDTFSTTYAGQRIAT 89
G D IV+ IE RI+DFTFIP E+GE VLHYEVGQ+YEPH+DYF D F+ GQRIAT
Sbjct: 172 GHDEIVEEIENRISDFTFIPPENGEGLQVLHYEVGQRYEPHHDYFFDEFNVRKGGQRIAT 231
Query: 90 MLMYLSDVEEGGETVFPNAKGNFSSVPWWNELSDCGKGGLSIKPKMGNAILFWSMKPDAT 149
+LMYLSDV+EGGETVFP AKGN S VPWW+ELS CGK GLS+ PK +A+LFWSMKPDA+
Sbjct: 232 VLMYLSDVDEGGETVFPAAKGNVSDVPWWDELSQCGKEGLSVLPKKRDALLFWSMKPDAS 291
Query: 150 LDPSSLHG 157
LDPSSLHG
Sbjct: 292 LDPSSLHG 299
>UniRef100_Q84T04 Putative oxidoreductase [Oryza sativa]
Length = 299
Score = 199 bits (505), Expect = 4e-50
Identities = 93/181 (51%), Positives = 127/181 (69%), Gaps = 7/181 (3%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M KS V D ++G + S+ RTSSG FL + D IV IE+R+A +TF+P E+ E+ +LH
Sbjct: 69 MEKSMVADNDSGKSIMSQVRTSSGTFLSKHEDDIVSGIEKRVAAWTFLPEENAESIQILH 128
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKG---NFSSVPW 117
YE+GQKY+ H+DYF D + G R+AT+LMYL+DV++GGETVFPNA G W
Sbjct: 129 YELGQKYDAHFDYFHDKNNLKRGGHRVATVLMYLTDVKKGGETVFPNAAGRHLQLKDETW 188
Query: 118 WNELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGE 177
SDC + GL++KPK G+A+LF+S+ +AT DP+SLHG+CPVI+G+KW KW+HV
Sbjct: 189 ----SDCARSGLAVKPKKGDALLFFSLHVNATTDPASLHGSCPVIEGEKWSATKWIHVRS 244
Query: 178 F 178
F
Sbjct: 245 F 245
>UniRef100_Q9LSI6 Prolyl 4-hydroxylase alpha subunit-like protein [Arabidopsis
thaliana]
Length = 332
Score = 196 bits (497), Expect = 3e-49
Identities = 89/179 (49%), Positives = 129/179 (71%), Gaps = 1/179 (0%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ KS V D ++G V+S RTSSG FL + D IV N+E ++A +TF+P E+GE+ +LH
Sbjct: 104 LEKSMVADNDSGESVESEVRTSSGMFLSKRQDDIVSNVEAKLAAWTFLPEENGESMQILH 163
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPH+DYF D + G RIAT+LMYLS+VE+GGETVFP KG + + +
Sbjct: 164 YENGQKYEPHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLK-DDS 222
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
++C K G ++KP+ G+A+LF+++ P+AT D +SLHG+CPV++G+KW +W+HV F+
Sbjct: 223 WTECAKQGYAVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWIHVKSFE 281
>UniRef100_Q8VZD7 AT3g28480/MFJ20_16 [Arabidopsis thaliana]
Length = 316
Score = 196 bits (497), Expect = 3e-49
Identities = 89/179 (49%), Positives = 129/179 (71%), Gaps = 1/179 (0%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ KS V D ++G V+S RTSSG FL + D IV N+E ++A +TF+P E+GE+ +LH
Sbjct: 88 LEKSMVADNDSGESVESEVRTSSGMFLSKRQDDIVNNVEAKLAAWTFLPEENGESMQILH 147
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPH+DYF D + G RIAT+LMYLS+VE+GGETVFP KG + + +
Sbjct: 148 YENGQKYEPHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLK-DDS 206
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
++C K G ++KP+ G+A+LF+++ P+AT D +SLHG+CPV++G+KW +W+HV F+
Sbjct: 207 WTECAKQGYAVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWIHVKSFE 265
>UniRef100_Q8L970 Prolyl 4-hydroxylase, putative [Arabidopsis thaliana]
Length = 316
Score = 196 bits (497), Expect = 3e-49
Identities = 89/179 (49%), Positives = 129/179 (71%), Gaps = 1/179 (0%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ KS V D ++G V+S RTSSG FL + D IV N+E ++A +TF+P E+GE+ +LH
Sbjct: 88 LEKSMVADNDSGESVESEVRTSSGMFLSKRQDDIVSNVEAKLAAWTFLPEENGESMQILH 147
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPH+DYF D + G RIAT+LMYLS+VE+GGETVFP KG + + +
Sbjct: 148 YENGQKYEPHFDYFHDQANLELGGHRIATVLMYLSNVEKGGETVFPMWKGKATQLK-DDS 206
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
++C K G ++KP+ G+A+LF+++ P+AT D +SLHG+CPV++G+KW +W+HV F+
Sbjct: 207 WTECAKQGYAVKPRKGDALLFFNLHPNATTDSNSLHGSCPVVEGEKWSATRWIHVKSFE 265
>UniRef100_Q84T01 Putative oxidoreductase [Oryza sativa]
Length = 310
Score = 191 bits (486), Expect = 6e-48
Identities = 91/178 (51%), Positives = 122/178 (68%), Gaps = 1/178 (0%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +S V D E+G V S RTSSG FL + D +V IE RIA +T +P E+ EN +L
Sbjct: 80 LKRSMVADNESGKSVMSEVRTSSGMFLDKQQDPVVSGIEERIAAWTLLPQENAENIQILR 139
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKY+PH+DYF D + G R AT+L YLS VE+GGETVFPNA+G + S P +
Sbjct: 140 YENGQKYDPHFDYFQDKVNQLQGGHRYATVLTYLSTVEKGGETVFPNAEG-WESQPKDDS 198
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
SDC K GL++K G+++LF++++PD T DP SLHG+CPVI+G+KW KW+HV +
Sbjct: 199 FSDCAKKGLAVKAVKGDSVLFFNLQPDGTPDPLSLHGSCPVIEGEKWSAPKWIHVRSY 256
>UniRef100_Q8RWK0 Hypothetical protein At5g18900 [Arabidopsis thaliana]
Length = 298
Score = 190 bits (482), Expect = 2e-47
Identities = 92/180 (51%), Positives = 125/180 (69%), Gaps = 2/180 (1%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D ++G S RTSSG F+ +G D IV IE +I+ +TF+P E+GE+ VL
Sbjct: 69 LKRSAVADNDSGESKFSEVRTSSGTFISKGKDPIVSGIEDKISTWTFLPKENGEDIQVLR 128
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKY+ H+DYF D + G R+AT+LMYLS+V +GGETVFP+A+ V NE
Sbjct: 129 YEHGQKYDAHFDYFHDKVNIVRGGHRMATILMYLSNVTKGGETVFPDAEIPSRRVLSENE 188
Query: 121 --LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
LSDC K G+++KP+ G+A+LF+++ PDA DP SLHG CPVI+G+KW KW+HV F
Sbjct: 189 EDLSDCAKRGIAVKPRKGDALLFFNLHPDAIPDPLSLHGGCPVIEGEKWSATKWIHVDSF 248
>UniRef100_Q8LAN3 Prolyl 4-hydroxylase alpha subunit-like protein [Arabidopsis
thaliana]
Length = 298
Score = 189 bits (479), Expect = 4e-47
Identities = 91/180 (50%), Positives = 125/180 (68%), Gaps = 2/180 (1%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D ++G S RTSSG F+ +G D IV IE +I+ +TF+P E+GE+ VL
Sbjct: 69 LKRSAVADNDSGESKFSEVRTSSGTFISKGKDPIVSGIEDKISTWTFLPKENGEDIQVLR 128
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWN- 119
YE GQKY+ H+DYF D + G R+AT+LMYLS+V +GGETVFP+A+ V N
Sbjct: 129 YEHGQKYDAHFDYFHDKVNIVRGGHRMATILMYLSNVTKGGETVFPDAEIPSRRVLSENK 188
Query: 120 -ELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
+LSDC K G+++KP+ G+A+LF+++ PDA DP SLHG CPVI+G+KW KW+HV F
Sbjct: 189 EDLSDCAKRGIAVKPRKGDALLFFNLHPDAIPDPLSLHGGCPVIEGEKWSATKWIHVDSF 248
>UniRef100_Q8L8T9 Prolyl 4-hydroxylase alpha subunit-like protein [Arabidopsis
thaliana]
Length = 297
Score = 187 bits (476), Expect = 8e-47
Identities = 90/180 (50%), Positives = 124/180 (68%), Gaps = 2/180 (1%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D + G S RTSSG F+ +G D IV IE +++ +TF+P E+GE+ VL
Sbjct: 68 LQRSAVADNDNGESQVSDVRTSSGTFISKGKDPIVSGIEDKLSTWTFLPKENGEDLQVLR 127
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAK--GNFSSVPWW 118
YE GQKY+ H+DYF D + G RIAT+L+YLS+V +GGETVFP+A+ S
Sbjct: 128 YEHGQKYDAHFDYFHDKVNIARGGHRIATVLLYLSNVTKGGETVFPDAQEFSRRSLSENK 187
Query: 119 NELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
++LSDC K G+++KPK GNA+LF++++ DA DP SLHG CPVI+G+KW KW+HV F
Sbjct: 188 DDLSDCAKKGIAVKPKKGNALLFFNLQQDAIPDPFSLHGGCPVIEGEKWSATKWIHVDSF 247
>UniRef100_Q6ZL00 Prolyl 4-hydroxylase alpha-1 subunit-like protein [Oryza sativa]
Length = 313
Score = 187 bits (475), Expect = 1e-46
Identities = 89/179 (49%), Positives = 122/179 (67%), Gaps = 1/179 (0%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
M +S V D ++G V S RTSSG FL + D +V IE+RIA +TF+P E+ EN +L
Sbjct: 83 MQRSMVADNKSGKSVMSEVRTSSGMFLDKRQDPVVSRIEKRIAAWTFLPEENAENIQILR 142
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
YE GQKYEPH+DYF D + G R AT+LMYLS VE+GGETVFPNA+G + + P +
Sbjct: 143 YEHGQKYEPHFDYFHDKVNQALGGHRYATVLMYLSTVEKGGETVFPNAEG-WENQPKDDT 201
Query: 121 LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFK 179
S+C + GL++KP G+ +LF+S+ D DP SLHG+CPVI+G+KW KW+ + ++
Sbjct: 202 FSECAQKGLAVKPVKGDTVLFFSLHIDGVPDPLSLHGSCPVIEGEKWSAPKWIRIRSYE 260
>UniRef100_Q84W84 Putative prolyl 4-hydroxylase [Arabidopsis thaliana]
Length = 253
Score = 186 bits (471), Expect = 3e-46
Identities = 89/179 (49%), Positives = 124/179 (68%), Gaps = 2/179 (1%)
Query: 1 MHKSAVI-DEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVL 59
+ KS V+ D ++G DS RTSSG FL + D IV N+E ++A +TF+P E+GE +L
Sbjct: 29 LEKSMVVADVDSGESEDSEVRTSSGMFLTKRQDDIVANVEAKLAAWTFLPEENGEALQIL 88
Query: 60 HYEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWN 119
HYE GQKY+PH+DYF D + G RIAT+LMYLS+V +GGETVFPN KG + +
Sbjct: 89 HYENGQKYDPHFDYFYDKKALELGGHRIATVLMYLSNVTKGGETVFPNWKGKTPQLK-DD 147
Query: 120 ELSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
S C K G ++KP+ G+A+LF+++ + T DP+SLHG+CPVI+G+KW +W+HV F
Sbjct: 148 SWSKCAKQGYAVKPRKGDALLFFNLHLNGTTDPNSLHGSCPVIEGEKWSATRWIHVRSF 206
>UniRef100_Q9LSI5 Gb|AAF08583.1 [Arabidopsis thaliana]
Length = 328
Score = 185 bits (469), Expect = 5e-46
Identities = 87/173 (50%), Positives = 120/173 (69%), Gaps = 1/173 (0%)
Query: 6 VIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLHYEVGQ 65
V D ++G DS RTSSG FL + D IV N+E ++A +TF+P E+GE +LHYE GQ
Sbjct: 3 VADVDSGESEDSEVRTSSGMFLTKRQDDIVANVEAKLAAWTFLPEENGEALQILHYENGQ 62
Query: 66 KYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNELSDCG 125
KY+PH+DYF D + G RIAT+LMYLS+V +GGETVFPN KG + + S C
Sbjct: 63 KYDPHFDYFYDKKALELGGHRIATVLMYLSNVTKGGETVFPNWKGKTPQLK-DDSWSKCA 121
Query: 126 KGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEF 178
K G ++KP+ G+A+LF+++ + T DP+SLHG+CPVI+G+KW +W+HV F
Sbjct: 122 KQGYAVKPRKGDALLFFNLHLNGTTDPNSLHGSCPVIEGEKWSATRWIHVRSF 174
>UniRef100_Q6AVM9 Putative prolyl 4-hydroxylase alpha subunit [Oryza sativa]
Length = 319
Score = 184 bits (466), Expect = 1e-45
Identities = 92/181 (50%), Positives = 123/181 (67%), Gaps = 4/181 (2%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGSDRIVKNIERRIADFTFIPVEHGENFNVLH 60
+ +SAV D +G S RTSSG F+++ D IV IE +IA +TF+P E+GE+ VL
Sbjct: 90 LKRSAVADNLSGKSELSDARTSSGTFIRKSQDPIVAGIEEKIAAWTFLPKENGEDIQVLR 149
Query: 61 YEVGQKYEPHYDYFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNE 120
Y+ G+KYE HYDYF D +T G RIAT+LMYL+DV EGGETVFP A+ F+ NE
Sbjct: 150 YKHGEKYERHYDYFSDNVNTLRGGHRIATVLMYLTDVAEGGETVFPLAE-EFTESGTNNE 208
Query: 121 ---LSDCGKGGLSIKPKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGE 177
LS+C K G+++KP+ G+A+LF+++ PDA+ D SLH CPVIKG+KW KW+ V
Sbjct: 209 DSTLSECAKKGVAVKPRKGDALLFFNLSPDASKDSLSLHAGCPVIKGEKWSATKWIRVAS 268
Query: 178 F 178
F
Sbjct: 269 F 269
>UniRef100_Q7XEP1 Hypothetical protein [Oryza sativa]
Length = 369
Score = 179 bits (453), Expect = 4e-44
Identities = 96/228 (42%), Positives = 124/228 (54%), Gaps = 49/228 (21%)
Query: 1 MHKSAVIDEETGNGVDSRERTSSGAFLKRGS----------------------------- 31
+ KS V D E+G V S RTSSG FL++
Sbjct: 77 LEKSMVADNESGKSVMSEVRTSSGMFLEKKQVVFFCSLREMKFDQLFPIYISIVCKALNH 136
Query: 32 -------------------DRIVKNIERRIADFTFIPVEHGENFNVLHYEVGQKYEPHYD 72
D +V IE RIA +TF+P ++GE+ +LHY+ G+KYEPHYD
Sbjct: 137 VVIFAIWTSILSNRPVLKLDEVVARIEERIAAWTFLPPDNGESIQILHYQNGEKYEPHYD 196
Query: 73 YFMDTFSTTYAGQRIATMLMYLSDVEEGGETVFPNAKGNFSSVPWWNELSDCGKGGLSIK 132
YF D + G RIAT+LMYLSDV +GGET+FP A+G P + SDC K G ++K
Sbjct: 197 YFHDKNNQALGGHRIATVLMYLSDVGKGGETIFPEAEGKLLQ-PKDDTWSDCAKNGYAVK 255
Query: 133 PKMGNAILFWSMKPDATLDPSSLHGACPVIKGDKWLCAKWMHVGEFKI 180
P G+A+LF+S+ PDAT D SLHG+CPVI+G KW KW+HV F I
Sbjct: 256 PVKGDALLFFSLHPDATTDSDSLHGSCPVIEGQKWSATKWIHVRSFDI 303
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.137 0.431
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 340,425,743
Number of Sequences: 2790947
Number of extensions: 14595051
Number of successful extensions: 27462
Number of sequences better than 10.0: 286
Number of HSP's better than 10.0 without gapping: 224
Number of HSP's successfully gapped in prelim test: 62
Number of HSP's that attempted gapping in prelim test: 26812
Number of HSP's gapped (non-prelim): 303
length of query: 180
length of database: 848,049,833
effective HSP length: 119
effective length of query: 61
effective length of database: 515,927,140
effective search space: 31471555540
effective search space used: 31471555540
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC149547.9


