

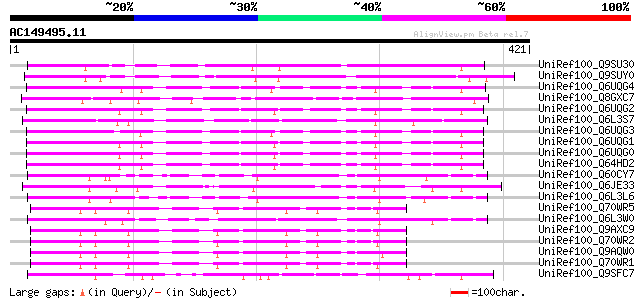
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149495.11 - phase: 0
(421 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis tha... 190 7e-47
UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis tha... 131 4e-29
UniRef100_Q6UQG4 S2 self-incompatibility locus-linked putative F... 122 2e-26
UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabid... 119 1e-25
UniRef100_Q6UQG2 S1 self-incompatibility locus-linked putative F... 119 2e-25
UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum] 117 6e-25
UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F... 115 2e-24
UniRef100_Q6UQG1 S2 self-incompatibility locus-linked putative F... 115 2e-24
UniRef100_Q6UQG0 S3 self-incompatibility locus-linked putative F... 113 1e-23
UniRef100_Q64HD2 S3 putative F-box protein SLF-S3B [Petunia hybr... 113 1e-23
UniRef100_Q60CY7 Putative F-box protein [Solanum demissum] 103 9e-21
UniRef100_Q6JE33 S3-locus linked F-box protein [Petunia integrif... 103 1e-20
UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum] 99 3e-19
UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum his... 99 3e-19
UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum] 98 4e-19
UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinu... 98 4e-19
UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum his... 98 4e-19
UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein)... 98 5e-19
UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum his... 98 5e-19
UniRef100_Q9SFC7 F17A17.21 protein [Arabidopsis thaliana] 97 1e-18
>UniRef100_Q9SU30 Hypothetical protein AT4g12560 [Arabidopsis thaliana]
Length = 408
Score = 190 bits (482), Expect = 7e-47
Identities = 123/391 (31%), Positives = 199/391 (50%), Gaps = 46/391 (11%)
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSL----NFKLILRHKT 70
+P DI+ +IF LP K+L+R R+ SK LI+ FI HL L + ++LR
Sbjct: 4 IPMDIVNDIFLRLPAKTLVRCRALSKPCYHLINDPDFIESHLHRVLQTGDHLMILLRGAL 63
Query: 71 NLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHPNDAT 130
LY +D +L S+ + HP + GS NGL+ + N P T
Sbjct: 64 RLYSVDLDSLD-SVSDVEHPMKRG-------GPTEVFGSSNGLIGLSNSP---------T 106
Query: 131 EITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLR-ISWLV 189
++ ++N +TR+ +P + +P + G +G G+D ++ DYK++R + + +
Sbjct: 107 DLAVFNPSTRQIHRLPPSSIDLP-----DGSSTRGYVFYGLGYDSVSDDYKVVRMVQFKI 161
Query: 190 SLQNSF---YHSHARLFSSKTNSWKIIPIMP----------YTVYYAQAMGVFVENSIHW 236
++ + ++FS K NSWK I + Y + Y + GV NS+HW
Sbjct: 162 DSEDELGCSFPYEVKVFSLKKNSWKRIESVASSIQLLFYFYYHLLYRRGYGVLAGNSLHW 221
Query: 237 IMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVN 296
++ + LIV F+L LE F+ V P + N N +++ VL GCL ++ N
Sbjct: 222 VLPRRPGLIAFNLIVRFDLALEEFEIVRFPEAV----ANGNVDIQMDIGVLDGCLCLMCN 277
Query: 297 YQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQIDCKKLV 356
Y + +DVW+MKEY RDSW K+FT+ K + +RPL YS D KVLL+++ KLV
Sbjct: 278 YDQSYVDVWMMKEYNVRDSWTKVFTVQKPKSVKSFSYMRPLVYSKDKKKVLLELNNTKLV 337
Query: 357 WYDLKSEQV--IYVEGIPNLDEAMICVESLV 385
W+DL+S+++ + ++ P+ A + V SLV
Sbjct: 338 WFDLESKKMSTLRIKDCPSSYSAELVVSSLV 368
>UniRef100_Q9SUY0 Hypothetical protein AT4g22390 [Arabidopsis thaliana]
Length = 394
Score = 131 bits (329), Expect = 4e-29
Identities = 114/422 (27%), Positives = 200/422 (47%), Gaps = 57/422 (13%)
Query: 13 SDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSL----NFKLILRH 68
++ P D++ E+F L +L++ R SK SLIDS +F++ HL+ L + ++LR
Sbjct: 2 AECPTDLINEMFLRLRATTLVKCRVLSKPCFSLIDSPEFVSSHLRRRLETGEHLMILLRG 61
Query: 69 KTNL--YQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTHP 126
L +LD P +++ + HP + GS NG++ + N P+
Sbjct: 62 PRLLRTVELDSP---ENVSDIPHPLQAG-------GFTEVFGSFNGVIGLCNSPV----- 106
Query: 127 NDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRIS 186
++ I+N +TRK +P P+ P +I R V +G G+D + D+K++RI
Sbjct: 107 ----DLAIFNPSTRKIHRLPIEPIDFPE----RDITREYV-FYGLGYDSVGDDFKVVRIV 157
Query: 187 WLVSLQNSFYHS---HARLFSSKTNSWKIIPIM----------PYTVYYAQAMGVFVENS 233
+ ++FS K NSWK + +M Y + + GV V N
Sbjct: 158 QCKLKEGKKKFPCPVEVKVFSLKKNSWKRVCLMFEFQILWISYYYHLLPRRGYGVVVNNH 217
Query: 234 IHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSM 293
+HWI+ + I+ ++L + + P + E+ ++++ VL GC+ +
Sbjct: 218 LHWILPRRQGVIAFNAIIKYDLASDDIGVLSFPQELYIED-------NMDIGVLDGCVCL 270
Query: 294 IVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQI-DC 352
+ + + +DVWV+KEY SW K++ + K ++ +RPL S D +K+LL+I +
Sbjct: 271 MCYDEYSHVDVWVLKEYEDYKSWTKLYRVPKPESVESVEFIRPLICSKDRSKILLEINNA 330
Query: 353 KKLVWYDLKSEQVIYVEGI--PNLDEAMICVESLV--PPSFPVD-NSSKKENRLSKSKRR 407
L+W+DL+S Q + GI + A I V SLV P SK + + KS +R
Sbjct: 331 ANLMWFDLES-QSLTTAGIECDSSFTADILVSSLVLGCKGDPTQAQRSKDQKMMPKSTKR 389
Query: 408 YF 409
++
Sbjct: 390 WY 391
>UniRef100_Q6UQG4 S2 self-incompatibility locus-linked putative F-box protein S2-A113
[Petunia integrifolia subsp. inflata]
Length = 376
Score = 122 bits (305), Expect = 2e-26
Identities = 113/400 (28%), Positives = 183/400 (45%), Gaps = 66/400 (16%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ I LPVKSLLRF+ + K+ ++I S F+NLHL + K L +
Sbjct: 3 ELPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFVNLHLNHTTKVKDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPLNH--------PFTTNIDPFTLNSIKA-----LIGSCNGLLAIYNGP 120
+ D N KSI+ P + +++ L + A LIG CNGL+A+
Sbjct: 63 KTDEYNFYKSILSFFSSKEDYDFMPMSPDVEIPHLTTTSARVFHQLIGPCNGLIAL---- 118
Query: 121 IAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDY 180
D+ ++N TRK+ +IP P IP R + GFGFD DY
Sbjct: 119 ------TDSLTTIVFNPATRKYRLIPPCPFGIPRGF------RRSISGIGFGFDSDVNDY 166
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWK-----IIPIMPYTVYYAQAMGVFVENSIH 235
K++R+S V + ++ +SW+ +PI VY+ + + + H
Sbjct: 167 KVVRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQEVPI----VYWLPCADILFKRNFH 221
Query: 236 WIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV 295
W +D +I+ F++ E F + +P ++ + + L +L C+S+I
Sbjct: 222 WFA-----FADDVVILCFDMNTEKFHNMGMPDAC---HFDDGKCYGL--VILCKCMSLIC 271
Query: 296 ------NYQTTKI-DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLL 348
+ T K+ D+W+MKEYG ++SW K R ++RL PL +D +LL
Sbjct: 272 YPDPMPSSPTEKLTDIWIMKEYGEKESWIK-------RCSIRLLPESPLAVWND-EILLL 323
Query: 349 QIDCKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESLVP 386
Q L+ YD S++V + + G+P +I ESL P
Sbjct: 324 QSKMGHLIAYDHNSDEVKELDLHGLPTSLRVIIYRESLTP 363
>UniRef100_Q8GXC7 Hypothetical protein At3g06240/F28L1_18 [Arabidopsis thaliana]
Length = 427
Score = 119 bits (299), Expect = 1e-25
Identities = 117/424 (27%), Positives = 190/424 (44%), Gaps = 78/424 (18%)
Query: 10 PVASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLK--------SSLN 61
P + LPP+I+ EI LP KS+ RFR SK +L F +HL SL+
Sbjct: 31 PESLVLPPEIITEILLRLPAKSIGRFRCVSKLFCTLSSDPGFAKIHLDLILRNESVRSLH 90
Query: 62 FKLILRHKTNLYQLDFPNL---TKSIIPLNHPFTTNIDPFTLNSI--------------- 103
KLI+ NLY LDF ++ + + + H + DP + +
Sbjct: 91 RKLIV-SSHNLYSLDFNSIGDGIRDLAAVEHNYPLKDDPSIFSEMIRNYVGDHLYDDRRV 149
Query: 104 -------------KALIGSCNGLLAIYNGPIAFTHPNDATEITIWNTNTRKHWIIP--FL 148
++GS NGL+ I G A + ++N T +P F
Sbjct: 150 MLKLNAKSYRRNWVEIVGSSNGLVCISPGEGA---------VFLYNPTTGDSKRLPENFR 200
Query: 149 PLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRISWLVSLQNSFYHSHARLFSSKTN 208
P +S ER +GFGFD LT DYKL++ LV+ A ++S K +
Sbjct: 201 P-------KSVEYERDNFQTYGFGFDGLTDDYKLVK---LVATSEDIL--DASVYSLKAD 248
Query: 209 SWKIIPIMPYTVY-YAQAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPA 267
SW+ I + Y + GV +IHW+ ++ ++VAF++ E F+E+P+P
Sbjct: 249 SWRRICNLNYEHNDGSYTSGVHFNGAIHWVFTESRHNQR--VVVAFDIQTEEFREMPVPD 306
Query: 268 VIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRF 327
E+ ++ S + V L G L ++ + D+WVM EYG SW +I R
Sbjct: 307 --EAEDCSHRFS-NFVVGSLNGRLCVVNSCYDVHDDIWVMSEYGEAKSWSRI------RI 357
Query: 328 TLRLKSLRPLGYSSDGNKVLLQIDCKKLVW-YDLKSEQVIYVEGIPNLD--EAMICVESL 384
L +S++PL + + +VLL++D +++ ++ + + + G+ D EA VESL
Sbjct: 358 NLLYRSMKPLCSTKNDEEVLLELDGDLVLYNFETNASSNLGICGVKLSDGFEANTYVESL 417
Query: 385 VPPS 388
+ P+
Sbjct: 418 ISPN 421
>UniRef100_Q6UQG2 S1 self-incompatibility locus-linked putative F-box protein S1-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 119 bits (297), Expect = 2e-25
Identities = 112/395 (28%), Positives = 180/395 (45%), Gaps = 60/395 (15%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ IF LPVKSLLRF+ T K+ +I S FINLHL + NF L +
Sbjct: 3 ELPQDVVIYIFVMLPVKSLLRFKCTCKTFYHIIKSSTFINLHLNHTTNFNDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPL--------NHPFTTNIDPFTLNSIKA-----LIGSCNGLLAIYNGP 120
+ D N KSI+ P + +++ L + A LIG CNGL+ +
Sbjct: 63 ETDEYNFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACICHRLIGPCNGLIVL---- 118
Query: 121 IAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDY 180
D+ ++N T K+ +IP P IP R + GFGFD DY
Sbjct: 119 ------TDSLTTIVFNPATLKYRLIPPCPFGIPRGF------RRSISGIGFGFDSDANDY 166
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKII--PIMPYTVYYAQAMGVFVENSIHWIM 238
K++R+S V + ++ +SW+ + +P+ V++ + + + HW
Sbjct: 167 KVVRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPF-VFWFPCAEILYKRNFHWFA 224
Query: 239 EKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV--- 295
+D +I+ F++ E F + +P ++ +S+ L +L C+++I
Sbjct: 225 -----FADDVVILCFDMNTEKFHNMGMPDAC---HFDDGKSYGL--VILFKCMTLICYPD 274
Query: 296 ---NYQTTKI-DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQID 351
+ T K+ D+W+MKEYG ++SW K R ++RL PL D +LL
Sbjct: 275 PMPSSPTEKLTDIWIMKEYGEKESWIK-------RCSIRLLPESPLAVWKD-EILLLHSK 326
Query: 352 CKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESL 384
L+ YDL S +V + + G P +I ESL
Sbjct: 327 MGHLIAYDLNSNEVQELDLHGYPESLRIIIYRESL 361
>UniRef100_Q6L3S7 Putative F-Box protein [Solanum demissum]
Length = 372
Score = 117 bits (293), Expect = 6e-25
Identities = 108/390 (27%), Positives = 175/390 (44%), Gaps = 39/390 (10%)
Query: 11 VASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKT 70
+ S LP +I+ EI +P KSLL+F SK+ LI S KFI HL+ N K H+
Sbjct: 5 IISVLPHEIIIEILLKVPPKSLLKFMCVSKTWLELISSAKFIKTHLELIANDKEYSHHRI 64
Query: 71 NLYQLDFPNLTKSIIP--LNHPFTTNI----DPFTLNSIKA-LIGSCNGLLAIYNGPIAF 123
++Q N +P LN +T + P +I ++GS NGL+ +Y
Sbjct: 65 -IFQESACNFKVCCLPSMLNKERSTELFDIGSPMENPTIYTWIVGSVNGLICLY------ 117
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLL 183
+ E +WN +K +P L + N +GFG+D DYK++
Sbjct: 118 ---SKIEETVLWNPAVKKSKKLPTLGAKLRNGCSY-------YLKYGFGYDETRDDYKVV 167
Query: 184 RISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLD 243
I + S S ++S K +SW+ I + + G FV I+W + ++D
Sbjct: 168 VIQCIYEDSGS-CDSVVNIYSLKADSWRTINKFQGN-FLVNSPGKFVNGKIYWALSADVD 225
Query: 244 GSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV--NYQTTK 301
++C I++ +L E ++ + LP + S+ L + V+G LS++ + T
Sbjct: 226 TFNMCNIISLDLADETWRRLELP------DSYGKGSYPLALGVVGSHLSVLCLNCIEGTN 279
Query: 302 IDVWVMKEYGSRDSWCKIFTLVKSR----FTLRLKSLRPLGYSSDGNKVLLQIDCKKLVW 357
DVW+ K+ G SW KIFT+ + F Y S+ +++LL + ++
Sbjct: 280 SDVWIRKDCGVEVSWTKIFTVDHPKDLGEFIFFTSIFSVPCYQSNKDEILLLLP-PVILT 338
Query: 358 YDLKSEQVIYVEGIPNLDEAMICVESLVPP 387
Y+ + QV V+ A I VESLV P
Sbjct: 339 YNGSTRQVEVVDRFEECAAAEIYVESLVDP 368
>UniRef100_Q6UQG3 S3 self-incompatibility locus-linked putative F-box protein S3-
A113 [Petunia integrifolia subsp. inflata]
Length = 376
Score = 115 bits (288), Expect = 2e-24
Identities = 112/402 (27%), Positives = 180/402 (43%), Gaps = 74/402 (18%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ I LPVKSLLRF+ + K+ ++I S FI LHL + N K L +
Sbjct: 3 ELPQDVVIYILVMLPVKSLLRFKCSCKTFCNIIKSSTFIYLHLNHTTNVKDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPLNHPFTTNIDPFTLNSIK-----------------ALIGSCNGLLAI 116
+ D N KSI+ F ++ + + SI LIG CNGL+A+
Sbjct: 63 KTDDYNFYKSIL----SFLSSKEGYDFKSISPDVEIPHLTTTSACVFHQLIGPCNGLIAL 118
Query: 117 YNGPIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPL 176
D+ ++N TRK+ +IP P IP R + GFGFD
Sbjct: 119 ----------TDSLTTIVFNPATRKYRLIPPCPFGIPRGF------RRSISGIGFGFDSD 162
Query: 177 TGDYKLLRISWLVSLQNSFYHSHARLFSSKTNSWK-----IIPIMPYTVYYAQAMGVFVE 231
DYK++R+S V + ++ +SW+ +PI VY+ + +
Sbjct: 163 ANDYKVVRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQEVPI----VYWLPCAEILYK 217
Query: 232 NSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCL 291
+ HW +D +I+ F++ E F + +P ++ + + L +L C+
Sbjct: 218 RNFHWFA-----FADDVVILCFDMNTEKFHNLGMPDAC---HFDDGKCYGL--VILCKCM 267
Query: 292 SMIV------NYQTTKI-DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGN 344
++I + T K+ D+W+MKEYG ++SW K R ++RL PL D
Sbjct: 268 TLICYPDPMPSSPTEKLTDIWIMKEYGEKESWIK-------RCSIRLLPESPLAVWMD-E 319
Query: 345 KVLLQIDCKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESL 384
+LLQ L+ YD S++V + + G+P +I ESL
Sbjct: 320 ILLLQSKIGHLIAYDHNSDEVKELDLHGLPTSLRVIIYRESL 361
>UniRef100_Q6UQG1 S2 self-incompatibility locus-linked putative F-box protein S2-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 115 bits (288), Expect = 2e-24
Identities = 110/395 (27%), Positives = 179/395 (44%), Gaps = 60/395 (15%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ IF LPVKSLLRF+ T K+ +I S FINLHL + NF L +
Sbjct: 3 ELPQDVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPL--------NHPFTTNIDPFTLNSIKA-----LIGSCNGLLAIYNGP 120
+ D KSI+ P + +++ L + A LIG CNGL+ +
Sbjct: 63 ETDEYKFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVL---- 118
Query: 121 IAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDY 180
D+ ++N T K+ +IP P IP R + GFGFD DY
Sbjct: 119 ------TDSLTTIVFNPATLKYRLIPPCPFGIPRGF------RRSISGIGFGFDSDANDY 166
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKII--PIMPYTVYYAQAMGVFVENSIHWIM 238
K++R+S V + ++ +SW+ + +P+ V++ + + + HW
Sbjct: 167 KVVRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPF-VFWFPCAEILYKRNFHWFA 224
Query: 239 EKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV--- 295
+D+ +I+ F++ E F + +P ++ + + L +L C+++I
Sbjct: 225 -----FADVVVILCFDMNTEKFHNMGMPDAC---HFDDGKCYGL--VILFKCMTLICYPD 274
Query: 296 ---NYQTTKI-DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQID 351
+ T K+ D+W+MKEYG ++SW K R ++RL PL D +LL
Sbjct: 275 PKPSSPTEKLTDIWIMKEYGEKESWMK-------RCSIRLLPESPLAVWKD-EILLLHSK 326
Query: 352 CKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESL 384
L+ YDL S +V + + G P +I ESL
Sbjct: 327 MGHLMAYDLNSNEVQELDLHGYPESLRIIIYRESL 361
>UniRef100_Q6UQG0 S3 self-incompatibility locus-linked putative F-box protein S3-
A134 [Petunia integrifolia subsp. inflata]
Length = 379
Score = 113 bits (282), Expect = 1e-23
Identities = 111/395 (28%), Positives = 177/395 (44%), Gaps = 60/395 (15%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ IF LPVKSLLR + T K+ +I S FINLHL + NF L +
Sbjct: 3 ELPQDVVIYIFVMLPVKSLLRSKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPL--------NHPFTTNIDPFTLNSIKA-----LIGSCNGLLAIYNGP 120
+ D N KSI+ P + ++ L + A LIG CNGL+ +
Sbjct: 63 ETDEYNFYKSILSFLFAKKDYDFKPISPDVKIPHLTTTAACICHRLIGPCNGLIVL---- 118
Query: 121 IAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDY 180
D+ ++N T K+ +IP P IP R + GFGFD DY
Sbjct: 119 ------TDSLTTIVFNPATLKYRLIPPCPFGIPRGF------RRSISGIGFGFDSDANDY 166
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKII--PIMPYTVYYAQAMGVFVENSIHWIM 238
K++R+S V ++ +SW+ + +P+ V++ + + + HW
Sbjct: 167 KVVRLS-EVYKGTCDKKMKVDIYDFSVDSWRELLGQDVPF-VFWFPCAEILYKRNFHWFA 224
Query: 239 EKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV--- 295
+D +I+ F++ E F + +P ++ +S+ L +L C+++I
Sbjct: 225 -----FADDVVILCFDMNTEKFHNMGMPDAC---HFDDGKSYGL--VILFKCMTLICYPD 274
Query: 296 ---NYQTTKI-DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQID 351
+ T K+ D+W+MKEYG ++SW K R ++RL PL D +LL
Sbjct: 275 PMPSSPTEKLTDIWIMKEYGEKESWIK-------RCSIRLLPESPLAVWKD-EILLLHSK 326
Query: 352 CKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESL 384
L+ YDL S +V + + G P +I ESL
Sbjct: 327 MGHLIAYDLNSNEVQELDLHGYPESLRIIIYRESL 361
>UniRef100_Q64HD2 S3 putative F-box protein SLF-S3B [Petunia hybrida]
Length = 379
Score = 113 bits (282), Expect = 1e-23
Identities = 108/395 (27%), Positives = 174/395 (43%), Gaps = 60/395 (15%)
Query: 14 DLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLY 73
+LP D++ IF LPVKSLLRF+ T K+ +I S FINLHL + NF L +
Sbjct: 3 ELPQDVVIYIFVMLPVKSLLRFKCTCKTFCHIIKSSTFINLHLNHTTNFNDELVLLKRSF 62
Query: 74 QLDFPNLTKSIIPL--------NHPFTTNIDPFTLNSIKA-----LIGSCNGLLAIYNGP 120
+ D N KSI+ P + +++ L + A LIG CNGL+ +
Sbjct: 63 ETDEYNFYKSILSFLFAKEDYDFKPISPDVEIPHLTTTAACVCHRLIGPCNGLIVL---- 118
Query: 121 IAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDY 180
D+ ++N T K+ +IP P IP R + GFGFD DY
Sbjct: 119 ------TDSLTTIVFNPATLKYRLIPPCPFGIPRGF------RRSISGIGFGFDSDANDY 166
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKII--PIMPYTVYYAQAMGVFVENSIHWIM 238
K++R+S V + ++ +SW+ + +P+ V++ + + + HW
Sbjct: 167 KVVRLS-EVYKEPCDKEMKVDIYDFSVDSWRELLGQDVPF-VFWFPCAEILYKRNFHWFA 224
Query: 239 EKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIV--- 295
+D+ +I+ F + E F + +P + + + L +L C+++I
Sbjct: 225 -----FADVVVILCFEMNTEKFHNMGMPDAC---HFADGKCYGL--VILFKCMTLICYPD 274
Query: 296 ----NYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSSDGNKVLLQID 351
+ D+W+MKEYG ++SW K R ++RL PL D +LL
Sbjct: 275 PMPSSPTEXWTDIWIMKEYGEKESWIK-------RCSIRLLPESPLAVWKD-EILLLHSK 326
Query: 352 CKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESL 384
L+ YD S +V + + G P +I ESL
Sbjct: 327 TGHLIAYDFNSNEVQELDLHGYPESLRIIIYRESL 361
>UniRef100_Q60CY7 Putative F-box protein [Solanum demissum]
Length = 383
Score = 103 bits (257), Expect = 9e-21
Identities = 106/395 (26%), Positives = 176/395 (43%), Gaps = 63/395 (15%)
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFK------LILRH 68
LP +++ EI LP+KSL +F SKS LI S F+ H+K + N K LI R+
Sbjct: 11 LPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKKHIKLTANDKGYIYHRLIFRN 70
Query: 69 KTNLYQLD-----FPN--LTKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPI 121
N ++ F N L + I+ ++ P I+ TL++ ++GS NGL+ +
Sbjct: 71 TNNDFKFCPLPPLFTNQQLIEEILHIDSP----IERTTLST--HIVGSVNGLICV----- 119
Query: 122 AFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGV-CIHGFGFDPLTGDY 180
H E IWN K +P + N+ G+ C GFG+D DY
Sbjct: 120 --AHVRQ-REAYIWNPAITKSKELP---------KSTSNLCSDGIKC--GFGYDESRDDY 165
Query: 181 KLLRISWLVSLQNSFYHSH---ARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWI 237
K++ I + + H+H ++S +TNSW + ++ G FV ++W
Sbjct: 166 KVVFIDYPIR------HNHRTVVNIYSLRTNSWTTLHDQLQGIFLLNLHGRFVNGKLYWT 219
Query: 238 MEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNY 297
++ +C I +F+L + + LP+ + S+ + V V+G LS++
Sbjct: 220 SSTCINNYKVCNITSFDLADGTWGSLELPS------CGKDNSY-INVGVVGSDLSLLYTC 272
Query: 298 Q--TTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRP---LGYSSDGNKVLLQIDC 352
Q DVW+MK G SW K+FT+ + + + P ++LL +D
Sbjct: 273 QLGAATSDVWIMKHSGVNVSWTKLFTIKYPQNIKIHRCVAPAFTFSIHIRHGEILLLLDS 332
Query: 353 KKLVWYDLKSEQVIYVEGIPNLDEAMICVESLVPP 387
++ YD + Q+ + + +E I VESLV P
Sbjct: 333 AIMI-YDGSTRQLKHTFHVNQCEE--IYVESLVNP 364
>UniRef100_Q6JE33 S3-locus linked F-box protein [Petunia integrifolia subsp. inflata]
Length = 388
Score = 103 bits (256), Expect = 1e-20
Identities = 108/420 (25%), Positives = 179/420 (41%), Gaps = 68/420 (16%)
Query: 11 VASDLPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFK---LILR 67
V LP D++ I PVKSL+RF+ SK+ LI S FIN H+ N K ++ +
Sbjct: 5 VLKKLPEDLVCLILLTFPVKSLMRFKCISKTWSILIQSTTFINRHVNRKTNTKDEFILFK 64
Query: 68 HKTNLYQLDFPNLTKSII---PLNHPFTTNIDPFTLNS-----IKALIGSCNGLLAIYNG 119
Q +F ++ + + +P +ID + S LIG C+GL+A+
Sbjct: 65 RAIKDEQEEFRDILSFLSGHDDVLNPLFADIDVSYMTSKCNCAFNPLIGPCDGLIAL--- 121
Query: 120 PIAFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGD 179
D I N TR ++P P P ++E GV GFG D ++
Sbjct: 122 -------TDTIITIILNPATRNFRLLPPSPFGSPKGYH-RSVE--GV---GFGLDTISNY 168
Query: 180 YKLLRISWLVSLQNSFY----HSHARLFSSKTNSWKIIP-IMPYTVYYAQAMGVFVENSI 234
YK++RIS + ++ Y S F T+SW+ + + +Y+ G+ + +
Sbjct: 169 YKVVRISEVYCEEDGGYPGPKDSKIDAFDLSTDSWRELDHVQLPLIYWLPCSGMLYKEMV 228
Query: 235 HWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
HW ++ LC F+++ E+F+ + +P V + Q + L +L ++I
Sbjct: 229 HWFATTDMSTVILC----FDMSTEMFRNMKMPDTC---SVTHKQYYGL--VILCESFTLI 279
Query: 295 --------VNYQTTKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSS----- 341
++ K+ +WVM EYG +SW +T +RPL S
Sbjct: 280 GYPNPVSPIDPAHDKMHIWVMMEYGVSESWIMKYT------------IRPLSIESPLAVW 327
Query: 342 DGNKVLLQIDCKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESLVPPSFPVDNSSKKEN 399
N +LLQ L+ YDL S Q + + G P+ ++ E L + S+K +N
Sbjct: 328 KKNILLLQSSSGLLISYDLNSGQAKELNLHGFPDSLSVIVYKECLTSIQNGSEYSTKVQN 387
>UniRef100_Q6L3L6 Putative F-box-like protein [Solanum demissum]
Length = 384
Score = 98.6 bits (244), Expect = 3e-19
Identities = 104/394 (26%), Positives = 175/394 (44%), Gaps = 61/394 (15%)
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFK------LILRH 68
LP +++ EI LP+KSL +F SKS LI S F+ H+K + N K LI R+
Sbjct: 12 LPDELITEILLRLPIKSLSKFMCVSKSWLQLISSPAFVKNHIKLTANGKGYIYHRLIFRN 71
Query: 69 KTNLYQL-DFPNL--TKSIIPLNHPFTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTH 125
+ ++ P+L + +I + I+ TL++ ++GS NGL I H
Sbjct: 72 TNDDFKFCPLPSLFTKQQLIEELFDIVSPIERTTLST--HIVGSVNGL-------ICAAH 122
Query: 126 PNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGV-CIHGFGFDPLTGDYKLLR 184
E IWN K +P N+ G+ C GFG+D DYK++
Sbjct: 123 VRQ-REAYIWNPTITKSKELP---------KSRSNLCSDGIKC--GFGYDESHDDYKVVF 170
Query: 185 ISWLVSLQNSFYHSH---ARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKN 241
I++ +H+H ++S +TNSW + ++ FV+ ++W
Sbjct: 171 INY------PSHHNHRSVVNIYSLRTNSWTTLHDQLQGIFLLNLHCRFVKEKLYWTSSTC 224
Query: 242 LDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQ--T 299
++ +C I +F+L ++ + LP+ + S+ + V V+G LS++ Q
Sbjct: 225 INNYKVCNITSFDLADGTWESLELPS------CGKDNSY-INVGVVGSDLSLLYTCQRGA 277
Query: 300 TKIDVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLR------PLGYSSDGNKVLLQIDCK 353
DVW+MK G SW K+FT+ ++ +K R ++LL +D
Sbjct: 278 ANSDVWIMKHSGVNVSWTKLFTI---KYPQNIKIHRCVVPAFTFSIHIRHGEILLVLDSA 334
Query: 354 KLVWYDLKSEQVIYVEGIPNLDEAMICVESLVPP 387
++ YD + Q+ + + +E I VESLV P
Sbjct: 335 IMI-YDGSTRQLKHTFHVNQCEE--IYVESLVNP 365
>UniRef100_Q70WR5 S locus F-box (SLF)-S4 protein [Antirrhinum hispanicum]
Length = 376
Score = 98.6 bits (244), Expect = 3e-19
Identities = 99/333 (29%), Positives = 151/333 (44%), Gaps = 61/333 (18%)
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL---KSSLNFKLILRH------ 68
D+ +EI VKSLLRFR SKS SLI SH FI+ HL +++ N ++ R+
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSHDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 69 -KTNLYQLDFPNLTKSIIPLNHPFTTNI----DPFTLNSIKALIGSCNGLLAIYNGPIAF 123
+ Y ++ P L + + L +P+ NI D F L L+G CNGL+ + G
Sbjct: 70 DMFSFYDINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLVCLAYGDC-- 127
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC--IHGFGFDPLTGD-Y 180
+ + N R+ +P P P G C I G+GF D Y
Sbjct: 128 --------VLLSNPALREIKRLPPTPFANPE----------GHCTDIIGYGFGNTCNDCY 169
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYA---QAMGVFVENSIHWI 237
K++ I S+ +H + ++ S TNSWK I + Y +F + + HW
Sbjct: 170 KVVLIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHW- 225
Query: 238 MEKNLDGSDLC---LIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
N + +D+ I+ F++ EVFKE+ P + + SF L + L CL+M
Sbjct: 226 ---NANSTDIFYADFILTFDIITEVFKEMAYPHCL----AQFSNSF-LSLMSLNECLAM- 276
Query: 295 VNY-----QTTKIDVWVMKEYGSRDSWCKIFTL 322
V Y + D+WVM +YG R+SW K + +
Sbjct: 277 VRYKEWMEEPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q6L3W0 Putative F-box protein [Solanum demissum]
Length = 427
Score = 98.2 bits (243), Expect = 4e-19
Identities = 98/388 (25%), Positives = 169/388 (43%), Gaps = 47/388 (12%)
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRHKTNLYQ 74
LP +++ EI LPVKSL +F SKS LI S F+ H+K + + K + H+
Sbjct: 53 LPDELITEILLKLPVKSLSKFMCVSKSWLQLISSPTFVKNHIKLTADDKGYIHHRLIFRN 112
Query: 75 LD----FPNLTKSIIPLNHP-----FTTNIDPFTLNSIKALIGSCNGLLAIYNGPIAFTH 125
+D F +L H + I+ TL++ ++GS NGL+ + +G
Sbjct: 113 IDGNFKFCSLPPLFTKQQHTEELFHIDSPIERSTLST--HIVGSVNGLICVVHG------ 164
Query: 126 PNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYKLLRI 185
E IWN K +P + + S +I+ +GFG+D DYK++ I
Sbjct: 165 ---QKEAYIWNPTITKSKELP----KFTSNMCSSSIK------YGFGYDESRDDYKVVFI 211
Query: 186 SWLVS-LQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYAQAMGVFVENSIHWIMEKNLDG 244
+ + +S + ++S + NSW + G FV ++W ++
Sbjct: 212 HYPYNHSSSSNMTTVVHIYSLRNNSWTTFRDQ-LQCFLVNHYGRFVNGKLYWTSSTCINK 270
Query: 245 SDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMIVNYQ--TTKI 302
+C I +F+L + + LP + +FD+ + V+G LS++ Q
Sbjct: 271 YKVCNITSFDLADGTWGSLDLP-------ICGKDNFDINLGVVGSDLSLLYTCQRGAATS 323
Query: 303 DVWVMKEYGSRDSWCKIFTLVKSRFTLRLKSLRPLGYSS---DGNKVLLQIDCKKLVWYD 359
DVW+MK SW K+FT+ + + P+ S +++LL + ++ YD
Sbjct: 324 DVWIMKHSAVNVSWTKLFTIKYPQNIKTHRCFAPVFTFSIHFRHSEILLLLRSAIMI-YD 382
Query: 360 LKSEQVIYVEGIPNLDEAMICVESLVPP 387
+ Q+ + + +E I VESLV P
Sbjct: 383 GSTRQLKHTSDVMQCEE--IYVESLVNP 408
>UniRef100_Q9AXC9 S locus F-box (SLF)-S2-like protein [Antirrhinum hispanicum]
Length = 376
Score = 98.2 bits (243), Expect = 4e-19
Identities = 99/333 (29%), Positives = 152/333 (44%), Gaps = 61/333 (18%)
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL---KSSLNFKLILRH------ 68
D+ +EI VKSLLRFR SKS SLI S+ FI+ HL +++ N ++ R+
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 69 -KTNLYQLDFPNLTKSIIPLNHPFTTNI----DPFTLNSIKALIGSCNGLLAIYNGPIAF 123
+ Y ++ P L + + L +P+ NI D F L L+G CNGL+ + G
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGDC-- 127
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC--IHGFGFDPLTGD-Y 180
+ + N R+ +P P P G C I G+GF D Y
Sbjct: 128 --------VLLSNPALREIKRLPPTPFANPE----------GHCTDIIGYGFGNTCNDCY 169
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYA---QAMGVFVENSIHWI 237
K++ I S+ +H + ++ S TNSWK I + Y +F + + HW
Sbjct: 170 KVVLIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHW- 225
Query: 238 MEKNLDGSDLC---LIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
N + +D+ I+ F++ EVFKE+ P + + SF L + L CL+M
Sbjct: 226 ---NANSTDIFYADFILTFDIITEVFKEMAYPHCL----AQFSNSF-LSLMSLNECLAM- 276
Query: 295 VNY-----QTTKIDVWVMKEYGSRDSWCKIFTL 322
V Y + D+WVMK+YG R+SW K + +
Sbjct: 277 VRYKEWMEEPELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q70WR2 S locus F-box (SLF)-S1 protein [Antirrhinum hispanicum]
Length = 376
Score = 98.2 bits (243), Expect = 4e-19
Identities = 99/333 (29%), Positives = 152/333 (44%), Gaps = 61/333 (18%)
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL---KSSLNFKLILRH------ 68
D+ +EI VKSLLRFR SKS SLI S+ FI+ HL +++ N ++ R+
Sbjct: 10 DVTSEILLFSSVKSLLRFRLVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 69 -KTNLYQLDFPNLTKSIIPLNHPFTTNI----DPFTLNSIKALIGSCNGLLAIYNGPIAF 123
+ Y ++ P L + + L +P+ NI D F L L+G CNGL+ + G
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGDC-- 127
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC--IHGFGFDPLTGD-Y 180
+ + N R+ +P P P G C I G+GF D Y
Sbjct: 128 --------VLLSNPALREIKRLPPTPFANPE----------GHCTDIIGYGFGNTCNDCY 169
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYA---QAMGVFVENSIHWI 237
K++ I S+ +H + ++ S TNSWK I + Y +F + + HW
Sbjct: 170 KVVLIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHW- 225
Query: 238 MEKNLDGSDLC---LIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
N + +D+ I+ F++ EVFKE+ P + + SF L + L CL+M
Sbjct: 226 ---NANSTDIFYADFILTFDIITEVFKEMAYPHCL----AQFSNSF-LSLMSLNECLAM- 276
Query: 295 VNY-----QTTKIDVWVMKEYGSRDSWCKIFTL 322
V Y + D+WVMK+YG R+SW K + +
Sbjct: 277 VRYKEWMEEPELFDIWVMKQYGVRESWTKQYVI 309
>UniRef100_Q9AQW0 SLF-S2 protein (S locus F-box (SLF)-S2 protein) [Antirrhinum
hispanicum]
Length = 376
Score = 97.8 bits (242), Expect = 5e-19
Identities = 96/332 (28%), Positives = 152/332 (44%), Gaps = 59/332 (17%)
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL---KSSLNFKLILRH------ 68
D+++EI VKSLLRFR SKS SLI S+ FI+ HL +++ N ++ R+
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 69 -KTNLYQLDFPNLTKSIIPLNHPFTTNI----DPFTLNSIKALIGSCNGLLAIYNGPIAF 123
+ Y ++ P L + + L +P+ NI D F L L+G CNGL+ + G
Sbjct: 70 DMFSFYNINSPELDELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGDC-- 127
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC--IHGFGFDPLTGD-Y 180
+ + N R+ +P P P G C I G+GF D Y
Sbjct: 128 --------VLLSNPALREIKRLPPTPFANPE----------GHCTDIIGYGFGNTCNDCY 169
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYA---QAMGVFVENSIHWI 237
K++ I S+ +H + ++ S TNSWK I + Y +F + + HW
Sbjct: 170 KVVLIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHW- 225
Query: 238 MEKNLDGSDLC---LIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
N + +D+ I+ F++ EVFKE+ P + + SF L + L CL+M+
Sbjct: 226 ---NANSTDIFYADFILTFDIITEVFKEMAYPHCL----AQFSNSF-LSLMSLNECLAMV 277
Query: 295 VNYQTTK----IDVWVMKEYGSRDSWCKIFTL 322
+ + D+WVM +YG R+SW K + +
Sbjct: 278 RYKEWMEDPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q70WR1 S locus F-box (SLF)-S5 protein [Antirrhinum hispanicum]
Length = 376
Score = 97.8 bits (242), Expect = 5e-19
Identities = 98/333 (29%), Positives = 152/333 (45%), Gaps = 61/333 (18%)
Query: 18 DILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHL---KSSLNFKLILRH------ 68
D+++EI VKSLLRFR SKS SLI S+ FI+ HL +++ N ++ R+
Sbjct: 10 DVISEILLFSSVKSLLRFRCVSKSWCSLIKSNDFIDNHLLRRQTNGNVMVVKRYVRTPER 69
Query: 69 -KTNLYQLDFPNLTKSIIPLNHPFTTNI----DPFTLNSIKALIGSCNGLLAIYNGPIAF 123
+ Y ++ P L + + L +P+ NI D F L L+G CNGL+ + G
Sbjct: 70 DMFSFYNINSPKLEELLPDLPNPYFKNIKFDYDYFYLPQRVNLMGPCNGLICLAYGDC-- 127
Query: 124 THPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVC--IHGFGFDPLTGD-Y 180
+ + N R+ +P P P G C I G+GF D Y
Sbjct: 128 --------VLLSNPALREIKRLPPTPFANPE----------GHCTDIIGYGFGNTCNDCY 169
Query: 181 KLLRISWLVSLQNSFYHSHARLFSSKTNSWKIIPIMPYTVYYA---QAMGVFVENSIHWI 237
K++ I S+ +H + ++ S TNSWK I + Y +F + + HW
Sbjct: 170 KVVLIE---SVGPEDHHINIYVYYSDTNSWKHIEDDSTPIKYICHFPCNELFFKGAFHW- 225
Query: 238 MEKNLDGSDLC---LIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLEVAVLGGCLSMI 294
N + +D+ I+ F++ EVFKE+ P + + SF L + L CL+M
Sbjct: 226 ---NANSTDIFYADFILTFDIITEVFKEMAYPHCL----AQFSNSF-LSLMSLNECLAM- 276
Query: 295 VNY-----QTTKIDVWVMKEYGSRDSWCKIFTL 322
V Y + D+WVM +YG R+SW K + +
Sbjct: 277 VRYKEWMEEPELFDIWVMNQYGVRESWTKQYVI 309
>UniRef100_Q9SFC7 F17A17.21 protein [Arabidopsis thaliana]
Length = 417
Score = 96.7 bits (239), Expect = 1e-18
Identities = 113/427 (26%), Positives = 179/427 (41%), Gaps = 88/427 (20%)
Query: 15 LPPDILAEIFSCLPVKSLLRFRSTSKSLRSLIDSHKFINLHLKSSLNFKLILRH----KT 70
LP DI+A+IFS LP+ S+ R +S RS++ H ++ S L+L +
Sbjct: 28 LPEDIIADIFSRLPISSIARLMFVCRSWRSVLTQHGRLSSSSSSPTKPCLLLHCDSPIRN 87
Query: 71 NLYQLDFPNLTKSIIPLNHPFTTNIDPFTLNSIKAL-----IGSCNGLL----AIYNGPI 121
L+ LD K I FTL ++ +GSCNGLL ++YN +
Sbjct: 88 GLHFLDLSEEEKRI---------KTKKFTLRFASSMPEFDVVGSCNGLLCLSDSLYNDSL 138
Query: 122 AFTHPNDATEITIWNTNTRKHWIIPFLPLPIPNIVESENIERGGVCIHGFGFDPLTGDYK 181
+P + TN+ L LP E N + GFGF +T +YK
Sbjct: 139 YLYNP--------FTTNS--------LELP-----ECSNKYHDQELVFGFGFHEMTKEYK 177
Query: 182 LLRISWL---VSLQNSFYHSHARL-----------FSSKTN----SWKIIPIMPYTVYYA 223
+L+I + S N Y R+ SSKT SW+ + PY +
Sbjct: 178 VLKIVYFRGSSSNNNGIYRGRGRIQYKQSEVQILTLSSKTTDQSLSWRSLGKAPYK-FVK 236
Query: 224 QAMGVFVENSIHWIMEKNLDGSDLCLIVAFNLTLEVFKEVPLPAVIRGEEVNNNQSFDLE 283
++ V +H++ D V+F+L E FKE+P P G N++ +L+
Sbjct: 237 RSSEALVNGRLHFVTRPRRHVPDR-KFVSFDLEDEEFKEIPKPDC-GGLNRTNHRLVNLK 294
Query: 284 VAVLGGCLSMIVNYQTTKIDVWVMKEYGSRDSWCKIFTL-------VKSRFTLRL----- 331
GCL +V K+D+WVMK YG ++SW K +++ +K +
Sbjct: 295 -----GCLCAVVYGNYGKLDIWVMKTYGVKESWGKEYSIGTYLPKGLKQNLDRPMWIWKN 349
Query: 332 ----KSLRPLGYSSDGNKVLLQIDCKKLVWYDLKSEQV--IYVEGIPNLDEAMICVESLV 385
K +R L +G ++LL+ + LV YD K + + G+PN ++ +L
Sbjct: 350 AENGKVVRVLCLLENG-EILLEYKSRVLVAYDPKLGKFKDLLFHGLPNWFHTVVHAGTLS 408
Query: 386 PPSFPVD 392
P+D
Sbjct: 409 WFDTPLD 415
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.323 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 726,370,775
Number of Sequences: 2790947
Number of extensions: 31143479
Number of successful extensions: 77448
Number of sequences better than 10.0: 303
Number of HSP's better than 10.0 without gapping: 189
Number of HSP's successfully gapped in prelim test: 114
Number of HSP's that attempted gapping in prelim test: 76998
Number of HSP's gapped (non-prelim): 381
length of query: 421
length of database: 848,049,833
effective HSP length: 130
effective length of query: 291
effective length of database: 485,226,723
effective search space: 141200976393
effective search space used: 141200976393
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 76 (33.9 bits)
Medicago: description of AC149495.11


