

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149494.4 - phase: 0
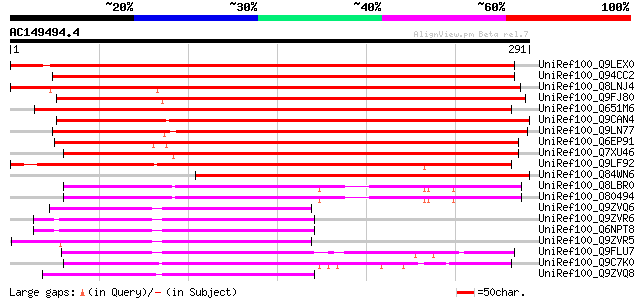
(291 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LEX0 Hypothetical protein T27I15_150 [Arabidopsis th... 361 1e-98
UniRef100_Q94CC2 Hypothetical protein At3g61060 [Arabidopsis tha... 353 2e-96
UniRef100_Q8LNJ4 Hypothetical protein OSJNBb0028C01.26 [Oryza sa... 344 2e-93
UniRef100_Q9FJ80 Similarity to unknown protein [Arabidopsis thal... 329 5e-89
UniRef100_Q651M6 Hypothetical protein OSJNBa0047P18.29 [Oryza sa... 325 1e-87
UniRef100_Q9CAN4 Hypothetical protein F16M19.16 [Arabidopsis tha... 322 7e-87
UniRef100_Q9LN77 T12C24.23 [Arabidopsis thaliana] 320 3e-86
UniRef100_Q6EP91 F-box protein-like [Oryza sativa] 320 4e-86
UniRef100_Q7XU46 OSJNBa0088I22.7 protein [Oryza sativa] 313 3e-84
UniRef100_Q9LF92 Hypothetical protein F8J2_170 [Arabidopsis thal... 290 3e-77
UniRef100_Q84WN6 Hypothetical protein At1g63090 [Arabidopsis tha... 242 1e-62
UniRef100_Q8LBR0 Phloem-specific lectin PP2-like protein [Arabid... 103 7e-21
UniRef100_O80494 T12M4.17 protein [Arabidopsis thaliana] 102 1e-20
UniRef100_Q9ZVQ6 Putative phloem-specific lectin [Arabidopsis th... 99 2e-19
UniRef100_Q9ZVR6 Putative phloem-specific lectin [Arabidopsis th... 92 2e-17
UniRef100_Q6NPT8 At2g02230 [Arabidopsis thaliana] 92 2e-17
UniRef100_Q9ZVR5 Lectin-like protein [Arabidopsis thaliana] 91 3e-17
UniRef100_Q9FLU7 Phloem-specific lectin-like protein [Arabidopsi... 91 4e-17
UniRef100_Q9C7K0 Hypothetical protein F14G9.14 [Arabidopsis thal... 91 4e-17
UniRef100_Q9ZVQ8 Putative phloem-specific lectin [Arabidopsis th... 87 7e-16
>UniRef100_Q9LEX0 Hypothetical protein T27I15_150 [Arabidopsis thaliana]
Length = 290
Score = 361 bits (926), Expect = 1e-98
Identities = 169/284 (59%), Positives = 222/284 (77%), Gaps = 4/284 (1%)
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MGA+IS + ++N++ S K L D+PE+C++ I+ LDP +IC+LAR+NR F R
Sbjct: 1 MGANISGGSPEFDRNDDVY---SRKLRLVDLPENCVALIMTRLDPPEICRLARLNRMFRR 57
Query: 61 ASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYS 120
ASSADF+WESKLP+ Y+ +A+K E L+ + KKD+Y KL QPN FD GTKE+ +DK +
Sbjct: 58 ASSADFIWESKLPANYRVIAHKVFDEITLTKLIKKDLYAKLSQPNLFDDGTKELWIDKNT 117
Query: 121 GQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSY 180
G++CL +SSK+L+ITGIDDRRYW +IPT+ESRF++ AY+QQ+WW EV GE E FP G+Y
Sbjct: 118 GRLCLSISSKALRITGIDDRRYWSHIPTDESRFQSAAYVQQIWWFEVGGEFEIQFPSGTY 177
Query: 181 SITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYL-HEPGQWAY 239
S+ FR+QLGK+SKRLGRR+CN + +HGWDIKPVRFQL+TSD Q+++S CYL + PG W++
Sbjct: 178 SLFFRIQLGKTSKRLGRRICNSEHIHGWDIKPVRFQLATSDNQQAVSLCYLNNNPGSWSH 237
Query: 240 YHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTE 283
YHVGDF VT P+ IKFS+ QIDCTHTKGGLCIDS +I P E
Sbjct: 238 YHVGDFKVTNPDVSTGIKFSMTQIDCTHTKGGLCIDSVLILPKE 281
>UniRef100_Q94CC2 Hypothetical protein At3g61060 [Arabidopsis thaliana]
Length = 269
Score = 353 bits (907), Expect = 2e-96
Identities = 161/260 (61%), Positives = 208/260 (79%), Gaps = 1/260 (0%)
Query: 25 KTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFL 84
K L D+PE+C++ I+ LDP +IC+LAR+NR F RASSADF+WESKLP+ Y+ +A+K
Sbjct: 1 KLRLVDLPENCVALIMTRLDPPEICRLARLNRMFRRASSADFIWESKLPANYRVIAHKVF 60
Query: 85 GEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWI 144
E L+ + KKD+Y KL QPN FD GTKE+ +DK +G++CL +SSK+L+ITGIDDRRYW
Sbjct: 61 DEITLTKLIKKDLYAKLSQPNLFDDGTKELWIDKNTGRLCLSISSKALRITGIDDRRYWS 120
Query: 145 YIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQ 204
+IPT+ESRF++ AY+QQ+WW EV GE E FP G+YS+ FR+QLGK+SKRLGRR+CN +
Sbjct: 121 HIPTDESRFQSAAYVQQIWWFEVGGEFEIQFPSGTYSLFFRIQLGKTSKRLGRRICNSEH 180
Query: 205 VHGWDIKPVRFQLSTSDGQRSISECYL-HEPGQWAYYHVGDFMVTKPNKPIKIKFSLAQI 263
+HGWD KPVRFQL+TSD Q+++S CYL + PG W++YHVGDF VT P+ IKFS+ QI
Sbjct: 181 IHGWDTKPVRFQLATSDNQQAVSLCYLNNNPGSWSHYHVGDFKVTNPDVSTGIKFSMTQI 240
Query: 264 DCTHTKGGLCIDSAIIRPTE 283
DCTHTKGGLCIDS +I P E
Sbjct: 241 DCTHTKGGLCIDSVLILPKE 260
>UniRef100_Q8LNJ4 Hypothetical protein OSJNBb0028C01.26 [Oryza sativa]
Length = 310
Score = 344 bits (882), Expect = 2e-93
Identities = 164/299 (54%), Positives = 213/299 (70%), Gaps = 13/299 (4%)
Query: 1 MGASISSTASIREKNNEESNS--SSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAF 58
MGA +S + E S + ++ GLGD+PE C + +L+ LD +IC+LAR+N AF
Sbjct: 1 MGAGVSGLFGLGGDEGETSAAVGGAAAAGLGDLPELCAAEVLLRLDAPEICRLARLNHAF 60
Query: 59 HRASSADFVWESKLPSCYKFLAN-----------KFLGEENLSTMSKKDVYTKLCQPNRF 107
A+ ADFVWE+KLP Y++L + + L KK++Y +L +P F
Sbjct: 61 RGAAGADFVWEAKLPENYRYLMSFVEGGGGGDDGRQLRRRRWRPAGKKEIYARLARPVPF 120
Query: 108 DGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEV 167
DGG+KE L+K G+VC+ +SSKSL ITGIDDRRYW +IPT ESRF +VAYLQQ+WW EV
Sbjct: 121 DGGSKEFWLEKSKGRVCMALSSKSLVITGIDDRRYWQHIPTAESRFYSVAYLQQIWWFEV 180
Query: 168 IGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSIS 227
+GE++F FPVG+YS+ FR+ LGK KR GRRVC+ + VHGWD KPVRFQLSTSDGQ S+S
Sbjct: 181 VGEIDFSFPVGTYSLYFRIHLGKFYKRFGRRVCSTEHVHGWDKKPVRFQLSTSDGQHSLS 240
Query: 228 ECYLHEPGQWAYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRE 286
+C L EPG W YH GDF+V+KP++ IK+KFS+AQIDCTHTKGGLC+DSA I P F++
Sbjct: 241 QCSLGEPGSWVLYHAGDFVVSKPDQTIKLKFSMAQIDCTHTKGGLCVDSAFIYPKGFQQ 299
>UniRef100_Q9FJ80 Similarity to unknown protein [Arabidopsis thaliana]
Length = 291
Score = 329 bits (844), Expect = 5e-89
Identities = 150/269 (55%), Positives = 203/269 (74%), Gaps = 6/269 (2%)
Query: 27 GLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFL-- 84
GL D+PE+CI+++ M+++P +IC LARVN++FHRAS +D VWE KLPS YKFL + L
Sbjct: 20 GLEDVPENCITAMFMYMEPPEICLLARVNKSFHRASRSDAVWEDKLPSNYKFLVRRILED 79
Query: 85 ----GEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDR 140
G ++ KK++Y +LC+PN FD GTKE LDK SG+V L +S K++KITGIDDR
Sbjct: 80 QQQVGVKDKLIYRKKEIYARLCRPNLFDTGTKEAWLDKRSGKVFLAISPKAMKITGIDDR 139
Query: 141 RYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVC 200
RYW +I ++ESRF ++ YL+Q+WW+E +G++ F F G YS+ F++QLGK ++ GR+ C
Sbjct: 140 RYWEHISSDESRFGSITYLRQIWWLEAVGKIRFEFAPGKYSLLFKIQLGKPIRKCGRKTC 199
Query: 201 NDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPNKPIKIKFSL 260
+ DQVHGWDIKPVRFQLSTSDGQ ++SE +L E G+W Y+H GDF+V N P+ +KFS+
Sbjct: 200 SLDQVHGWDIKPVRFQLSTSDGQCAMSERHLDESGRWVYHHAGDFVVENQNSPVWVKFSM 259
Query: 261 AQIDCTHTKGGLCIDSAIIRPTEFRERLK 289
QIDCTHTKGGLC+D II P E+R + K
Sbjct: 260 LQIDCTHTKGGLCLDCVIICPFEYRGKYK 288
>UniRef100_Q651M6 Hypothetical protein OSJNBa0047P18.29 [Oryza sativa]
Length = 287
Score = 325 bits (832), Expect = 1e-87
Identities = 152/269 (56%), Positives = 199/269 (73%), Gaps = 2/269 (0%)
Query: 15 NNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPS 74
++ +T LGD+PESC++ +L LDP +IC++AR++R F A+S D VWE+KLP
Sbjct: 6 SSSAGGGGGGETALGDLPESCVAEVLRRLDPPEICRMARLSRTFRGAASGDGVWEAKLPR 65
Query: 75 CY-KFLANKFLGEEN-LSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSL 132
Y + LA GE L + KK+VY +LC+ NR DGGTKE LDK G VC+ +SS++L
Sbjct: 66 NYARLLAVAADGEAAALEAIPKKEVYARLCRRNRLDGGTKEFWLDKGGGGVCMTISSRAL 125
Query: 133 KITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSS 192
ITGIDDRRYW +IP +ESRF VAYL Q+WW EV GE+EF FP G+YS+ FRL LG+
Sbjct: 126 SITGIDDRRYWNFIPNDESRFHAVAYLSQIWWFEVRGEVEFCFPEGTYSLFFRLHLGRPL 185
Query: 193 KRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPNK 252
KRLGRRV + + +HGWDIKPVRFQLSTSDGQ++ S+CYL +PG W +HVGDF+V N+
Sbjct: 186 KRLGRRVYSSEHIHGWDIKPVRFQLSTSDGQQAQSKCYLTDPGVWINHHVGDFVVKSSNE 245
Query: 253 PIKIKFSLAQIDCTHTKGGLCIDSAIIRP 281
+KI+F++ QIDCTHTKGGLC+DS ++P
Sbjct: 246 LVKIQFAMVQIDCTHTKGGLCVDSVAVKP 274
>UniRef100_Q9CAN4 Hypothetical protein F16M19.16 [Arabidopsis thaliana]
Length = 289
Score = 322 bits (825), Expect = 7e-87
Identities = 149/266 (56%), Positives = 199/266 (74%), Gaps = 2/266 (0%)
Query: 27 GLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGE 86
GLGD+PESC++ IL +LDP +IC+ +++N AFH AS ADFVWESKLP YK + K LG
Sbjct: 25 GLGDLPESCVALILQNLDPVEICRFSKLNTAFHGASWADFVWESKLPPDYKLILEKILGS 84
Query: 87 ENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYI 146
+ K+D++T L + N FD G K+ +DK +G +CL S+K L ITGIDDRRYW +I
Sbjct: 85 FP-DNLRKRDIFTFLSRVNSFDEGNKKAWVDKRTGGLCLCTSAKGLSITGIDDRRYWSHI 143
Query: 147 PTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVH 206
P+++SRF +VAY+QQ+WW +V GE++F FP G+YS+ FRLQLGK KR G +V + +QVH
Sbjct: 144 PSDDSRFASVAYVQQIWWFQVDGEIDFPFPAGTYSVYFRLQLGKPGKRFGWKVVDTEQVH 203
Query: 207 GWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTK-PNKPIKIKFSLAQIDC 265
GW+IKPVRFQLST DGQ S S+C L E G W++YH GDF+V K + KIKFS+ QIDC
Sbjct: 204 GWNIKPVRFQLSTEDGQHSSSQCMLTEAGNWSHYHAGDFVVGKSKSSSTKIKFSMTQIDC 263
Query: 266 THTKGGLCIDSAIIRPTEFRERLKKL 291
THTKGGLC+DS ++ P+ ++RL ++
Sbjct: 264 THTKGGLCVDSVVVYPSSCKDRLMQI 289
>UniRef100_Q9LN77 T12C24.23 [Arabidopsis thaliana]
Length = 291
Score = 320 bits (820), Expect = 3e-86
Identities = 156/269 (57%), Positives = 200/269 (73%), Gaps = 6/269 (2%)
Query: 25 KTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFL 84
K GLGD+PE+C++ I+ +LDP +IC+ +++NRAF AS AD VWESKLP Y+ + K L
Sbjct: 25 KPGLGDLPEACVAIIVENLDPVEICRFSKLNRAFRGASWADCVWESKLPQNYRDVLEKIL 84
Query: 85 G--EENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRY 142
G ENL K+ +Y L + N FD TK+V +DK + VCL +S+K L ITGIDDRRY
Sbjct: 85 GGFPENLQ---KRHLYAFLSRINSFDDATKKVWIDKRTSGVCLSISAKGLSITGIDDRRY 141
Query: 143 WIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCND 202
W +IPT+ESRF +VAYLQQ+WW EV GE++F FPVG+YSI FRLQLG+S K GRRVCN
Sbjct: 142 WSHIPTDESRFSSVAYLQQIWWFEVDGEIDFPFPVGTYSIFFRLQLGRSGKWFGRRVCNT 201
Query: 203 DQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPNK-PIKIKFSLA 261
+QVHGWDIKPVRFQL T DGQ S S+C L E G W +YH GD +V + N+ KIKFS+
Sbjct: 202 EQVHGWDIKPVRFQLWTEDGQYSSSQCMLTERGNWIHYHAGDVVVRESNRSSTKIKFSMT 261
Query: 262 QIDCTHTKGGLCIDSAIIRPTEFRERLKK 290
QIDCTHTKGGL +DS ++ P+ +++LK+
Sbjct: 262 QIDCTHTKGGLSLDSVVVYPSSCKDQLKR 290
>UniRef100_Q6EP91 F-box protein-like [Oryza sativa]
Length = 297
Score = 320 bits (819), Expect = 4e-86
Identities = 150/272 (55%), Positives = 199/272 (73%), Gaps = 12/272 (4%)
Query: 26 TGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFL------ 79
T LGD+PESC++++L++LDP +IC++AR+NRAF A+SAD VW KLP Y++L
Sbjct: 17 TSLGDMPESCVAAVLLYLDPPEICQVARLNRAFRGAASADCVWAGKLPVNYRYLLAFAAA 76
Query: 80 ANKFLGE------ENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLK 133
A+ G+ + S SKKD++ +LC+P FD G KE +DK G +CL +SSK++
Sbjct: 77 ADDEGGDGGHGNGKRSSPSSKKDIFARLCRPTPFDFGNKEFWIDKNKGGICLSISSKAMV 136
Query: 134 ITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSK 193
ITGIDDRRYW + TEESRF ++AYLQQ+WW+EV GEL+F FP GSYSI FRL LG+ +
Sbjct: 137 ITGIDDRRYWSQLATEESRFHHIAYLQQIWWLEVDGELDFCFPAGSYSIFFRLHLGRPYR 196
Query: 194 RLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPNKP 253
R+GRR+C +QVHGW+ KP RFQLSTSD Q + SE YL + G W YHVGDF+V ++
Sbjct: 197 RMGRRICGTEQVHGWEAKPTRFQLSTSDEQHATSEYYLEQEGSWILYHVGDFVVLNSDEL 256
Query: 254 IKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFR 285
+K+KFS+ QIDCTHTKGGLC+DS +I P +R
Sbjct: 257 MKLKFSMLQIDCTHTKGGLCVDSVLIYPKGYR 288
>UniRef100_Q7XU46 OSJNBa0088I22.7 protein [Oryza sativa]
Length = 279
Score = 313 bits (802), Expect = 3e-84
Identities = 145/271 (53%), Positives = 197/271 (72%), Gaps = 16/271 (5%)
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PESC++++L++LDP +ICK+AR+NRAF A+SAD VW +KLP+ Y++LA ++ S
Sbjct: 1 MPESCVAAVLLYLDPPEICKVARLNRAFRGAASADCVWAAKLPANYRYLAALAAAADDDS 60
Query: 91 ----------------TMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKI 134
M KK++Y +LC+P FDGGTKE ++K G +C+ +SSK++ I
Sbjct: 61 GGDGATEGNGSRCSSAAMIKKEIYARLCRPTPFDGGTKEFWMEKNKGGLCISISSKAMAI 120
Query: 135 TGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKR 194
TGIDDRRYW ++ TEESRF +VAYLQQ+WW+EV GE++F FP GSYS+ FRLQLG+ K
Sbjct: 121 TGIDDRRYWSHLSTEESRFHHVAYLQQIWWLEVAGEIDFCFPAGSYSLFFRLQLGRPHKY 180
Query: 195 LGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYLHEPGQWAYYHVGDFMVTKPNKPI 254
+GRRV + +HGW+IKP RFQLSTSD Q++ S+ YL+EPG W YHVGDF+V+ ++
Sbjct: 181 MGRRVYGYESIHGWNIKPTRFQLSTSDDQQATSQYYLNEPGNWILYHVGDFVVSSSDQLT 240
Query: 255 KIKFSLAQIDCTHTKGGLCIDSAIIRPTEFR 285
+KFS+ QIDCTHTKGGLC+DS I P R
Sbjct: 241 NLKFSMMQIDCTHTKGGLCVDSVFIYPKGHR 271
>UniRef100_Q9LF92 Hypothetical protein F8J2_170 [Arabidopsis thaliana]
Length = 300
Score = 290 bits (742), Expect = 3e-77
Identities = 143/292 (48%), Positives = 196/292 (66%), Gaps = 19/292 (6%)
Query: 1 MGASISSTASIREKNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHR 60
MG+S+S+ N+ +N + GLGDIPESC++ + M+L P +IC LA +NR+F
Sbjct: 1 MGSSLSNL-------NDGTNGLAMGPGLGDIPESCVACVFMYLTPPEICNLAGLNRSFRG 53
Query: 61 ASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLCQPNRFDGGTKEVLLDKYS 120
A+S+D VWE KLP Y+ L + L E ++SKKD++ L +P FD KEV +D+ +
Sbjct: 54 AASSDSVWEKKLPENYQDLLD-LLPPERYHSLSKKDIFAVLSRPIPFDDDNKEVWIDRVT 112
Query: 121 GQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGELEFVFPVGSY 180
G+VC+ +S++ + ITGI+DRRYW +IPTEESRF VAYLQQ+WW EV G + F P G Y
Sbjct: 113 GRVCMAISARGMSITGIEDRRYWNWIPTEESRFHVVAYLQQIWWFEVDGTVRFHLPPGVY 172
Query: 181 SITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQRSISECYL--------- 231
S++FR+ LG+ +KRLGRRVC+ + HGWD+KPVRF LSTSDGQ + E YL
Sbjct: 173 SLSFRIHLGRFTKRLGRRVCHFELTHGWDLKPVRFSLSTSDGQEASCEYYLDDVERNEAL 232
Query: 232 --HEPGQWAYYHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRP 281
H+ G W Y VG+F+V +I++S+ QIDCTH+KGGLC+DS I P
Sbjct: 233 GKHKRGYWIEYRVGEFIVNGSEPSTEIQWSMKQIDCTHSKGGLCVDSVFINP 284
>UniRef100_Q84WN6 Hypothetical protein At1g63090 [Arabidopsis thaliana]
Length = 188
Score = 242 bits (617), Expect = 1e-62
Identities = 109/188 (57%), Positives = 144/188 (75%), Gaps = 1/188 (0%)
Query: 105 NRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWW 164
N FD G K+ +DK +G +CL S+K L ITGIDDRRYW +IP+++SRF +VAY+QQ+WW
Sbjct: 1 NSFDEGNKKAWVDKRTGGLCLCTSAKGLSITGIDDRRYWSHIPSDDSRFASVAYVQQIWW 60
Query: 165 VEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGWDIKPVRFQLSTSDGQR 224
+V GE++F FP G+YS+ FRLQLGK KR G +V + +QVHGW+IKPVRFQLST DGQ
Sbjct: 61 FQVDGEIDFPFPAGTYSVYFRLQLGKPGKRFGWKVVDTEQVHGWNIKPVRFQLSTEDGQH 120
Query: 225 SISECYLHEPGQWAYYHVGDFMVTK-PNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTE 283
S S+C L E G W++YH GDF+V K + KIKFS+ QIDCTHTKGGLC+DS ++ P+
Sbjct: 121 SSSQCMLTEAGNWSHYHAGDFVVGKSKSSSTKIKFSMTQIDCTHTKGGLCVDSVVVYPSS 180
Query: 284 FRERLKKL 291
++RL ++
Sbjct: 181 CKDRLMQI 188
>UniRef100_Q8LBR0 Phloem-specific lectin PP2-like protein [Arabidopsis thaliana]
Length = 288
Score = 103 bits (256), Expect = 7e-21
Identities = 73/297 (24%), Positives = 125/297 (41%), Gaps = 54/297 (18%)
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PE+C+++IL P D A V+ F A +DFVWE LP+ Y + ++ +
Sbjct: 2 LPEACVATILSFTTPADTISSAAVSSVFRVAGDSDFVWEKFLPTDYCHVISRSTDPHRIF 61
Query: 91 TMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEE 150
+ SKK++Y LC+ D G K ++K SG++ +SS+ L IT D R YW + P +
Sbjct: 62 S-SKKELYRCLCESILIDNGRKIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWSPRSD 120
Query: 151 SRFKNVAYLQQMWWVEVIGELE--FVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGW 208
SRF L W+E+IG+++ + P +Y +++ + +G
Sbjct: 121 SRFSEXVQLIMTDWLEIIGKIQTGALSPNTNYGAYLIMKV-------------TSRAYGL 167
Query: 209 DIKPVRFQLSTSDGQRSISECYL------------------------HE----------- 233
D+ P + +G++ I YL HE
Sbjct: 168 DLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQMERVFYGQREQRMATHEVVRSHRREPEV 227
Query: 234 -PGQWAYYHVGDFMV--TKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRER 287
W +G+F + + ++ SL ++ KGG+ ID +RP + R
Sbjct: 228 RDDGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAIDGIEVRPKPLKVR 284
>UniRef100_O80494 T12M4.17 protein [Arabidopsis thaliana]
Length = 288
Score = 102 bits (254), Expect = 1e-20
Identities = 73/297 (24%), Positives = 125/297 (41%), Gaps = 54/297 (18%)
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PE+C+++IL P D A V+ F A +DFVWE LP+ Y + ++ +
Sbjct: 2 LPEACVATILSFTTPADTISSAAVSSVFRVAGDSDFVWEKFLPTDYCHVISRSTDPHRIF 61
Query: 91 TMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEE 150
+ SKK++Y LC+ D G K ++K SG++ +SS+ L IT D R YW + P +
Sbjct: 62 S-SKKELYRCLCESILIDNGRKIFKIEKLSGKISYILSSRDLSITWSDQRHYWSWSPRSD 120
Query: 151 SRFKNVAYLQQMWWVEVIGELE--FVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGW 208
SRF L W+E+IG+++ + P +Y +++ + +G
Sbjct: 121 SRFSEGVQLIMTDWLEIIGKIQTGALSPNTNYGAYLIMKV-------------TSRAYGL 167
Query: 209 DIKPVRFQLSTSDGQRSISECYL------------------------HE----------- 233
D+ P + +G++ I YL HE
Sbjct: 168 DLVPAETSIKVGNGEKKIKSTYLSCLDNKKQQMERVFYGQREQRMATHEVVRSHRREPEV 227
Query: 234 -PGQWAYYHVGDFMV--TKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRPTEFRER 287
W +G+F + + ++ SL ++ KGG+ ID +RP + R
Sbjct: 228 RDDGWMEIELGEFETGSGEGDDDKEVVMSLTEVKGYQLKGGIAIDGIEVRPKPLKVR 284
>UniRef100_Q9ZVQ6 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 272
Score = 98.6 bits (244), Expect = 2e-19
Identities = 55/148 (37%), Positives = 79/148 (53%), Gaps = 6/148 (4%)
Query: 23 SSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANK 82
S + PE CIS I+ +P D C A V++ F +D +WE LP+ Y+ L
Sbjct: 9 SESSPFDSFPEDCISYIISFTNPRDACVAATVSKTFESTVKSDIIWEKFLPADYESLI-- 66
Query: 83 FLGEENLSTMSKKDVYTKLCQ-PNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRR 141
+ SKK++Y LC P FD K V L+K SG+ CL +S+ +L I D+ +
Sbjct: 67 ---PPSRVFSSKKELYFSLCNDPVLFDDDKKSVWLEKASGKRCLMLSAMNLSIIWGDNPQ 123
Query: 142 YWIYIPTEESRFKNVAYLQQMWWVEVIG 169
YW +IP ESRF+ VA L+ + W E+ G
Sbjct: 124 YWQWIPIPESRFEKVAKLRDVCWFEIRG 151
>UniRef100_Q9ZVR6 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 317
Score = 92.0 bits (227), Expect = 2e-17
Identities = 52/159 (32%), Positives = 87/159 (54%), Gaps = 8/159 (5%)
Query: 14 KNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLP 73
+N+E S + +S+ +PE CIS ++ H P D C +A V+++ A+ +D VWE LP
Sbjct: 20 RNDEISVTRASR--FDALPEDCISKVISHTSPRDACVVASVSKSVKSAAQSDLVWEMFLP 77
Query: 74 SCYKFLANKFLGEENLSTMSKKDVYTKLCQPN-RFDGGTKEVLLDKYSGQVCLFMSSKSL 132
S Y L ++ + +SKK+++ L + + G K ++K SG+ C +S+ L
Sbjct: 78 SEYSSLV-----LQSANHLSKKEIFLSLADNSVLVENGKKSFWVEKASGKKCYMLSAMEL 132
Query: 133 KITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGEL 171
I D YW +I ES+F+ VA L+ + W EV G++
Sbjct: 133 TIIWGDSPAYWKWITVPESKFEKVAELRNVCWFEVRGKI 171
>UniRef100_Q6NPT8 At2g02230 [Arabidopsis thaliana]
Length = 336
Score = 92.0 bits (227), Expect = 2e-17
Identities = 52/159 (32%), Positives = 87/159 (54%), Gaps = 8/159 (5%)
Query: 14 KNNEESNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLP 73
+N+E S + +S+ +PE CIS ++ H P D C +A V+++ A+ +D VWE LP
Sbjct: 20 RNDEISVTRASR--FDALPEDCISKVISHTSPRDACVVASVSKSVKSAAQSDLVWEMFLP 77
Query: 74 SCYKFLANKFLGEENLSTMSKKDVYTKLCQPN-RFDGGTKEVLLDKYSGQVCLFMSSKSL 132
S Y L ++ + +SKK+++ L + + G K ++K SG+ C +S+ L
Sbjct: 78 SEYSSLV-----LQSANHLSKKEIFLSLADNSVLVENGKKSFWVEKASGKKCYMLSAMEL 132
Query: 133 KITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGEL 171
I D YW +I ES+F+ VA L+ + W EV G++
Sbjct: 133 TIIWGDSPAYWKWITVPESKFEKVAELRNVCWFEVRGKI 171
>UniRef100_Q9ZVR5 Lectin-like protein [Arabidopsis thaliana]
Length = 305
Score = 91.3 bits (225), Expect = 3e-17
Identities = 56/173 (32%), Positives = 90/173 (51%), Gaps = 10/173 (5%)
Query: 2 GASISSTASIREKNNEESNSSSSKTG---LGDIPESCISSILMHLDPHDICKLARVNRAF 58
G + ++S+ +K+ ES+ + +G ++PE CIS+I+ P D C A V++ F
Sbjct: 13 GRKVPGSSSMVQKHRVESSGGAIISGPSLFDNLPEDCISNIISFTSPRDACVAASVSKTF 72
Query: 59 HRASSADFVWESKLPSCYKFLANKFLGEENLSTMSKKDVYTKLC-QPNRFDGGTKEVLLD 117
A ++D VW+ LPS Y L + SKK++Y +C P + G K L+
Sbjct: 73 ESAVNSDSVWDKFLPSDYSSLV-----PPSRVFSSKKELYFAICDNPVLVEDGGKSFWLE 127
Query: 118 KYSGQVCLFMS-SKSLKITGIDDRRYWIYIPTEESRFKNVAYLQQMWWVEVIG 169
K +G+ C +S KS+ IT + +YW +I E+RF+ V L + W EV G
Sbjct: 128 KENGKKCFMLSPKKSMWITWVSTPQYWRWISIPEARFEEVPELLNVCWFEVRG 180
>UniRef100_Q9FLU7 Phloem-specific lectin-like protein [Arabidopsis thaliana]
Length = 251
Score = 90.9 bits (224), Expect = 4e-17
Identities = 62/264 (23%), Positives = 124/264 (46%), Gaps = 29/264 (10%)
Query: 30 DIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENL 89
D+PE CI++++ P D C+++ V++ A+ ++ WE LPS Y+ + +L
Sbjct: 5 DLPEECIATMISFTSPFDACRISAVSKLLRSAADSNTTWERFLPSDYRMYI-----DNSL 59
Query: 90 STMSKKDVYTKLCQ-PNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPT 148
S S K ++ + C+ P + G ++K SG+ C +S++ L I +D +WI++
Sbjct: 60 SRFSNKQLFLRFCESPLLIEDGRTSFWMEKRSGKKCWMLSARKLDIVWVDSPEFWIWVSI 119
Query: 149 EESRFKNVAYLQQMWWVEVIGELEFVFPVGSYSITFRLQLGKSSKRLGRRVCNDDQVHGW 208
+SRF+ VA L + W E+ G++ S S+ L K++ V + ++ +
Sbjct: 120 PDSRFEEVAGLLMVCWFEIRGKI-------STSL-----LSKATNYSAYLVFKEQEMGSF 167
Query: 209 DIKPVRFQLSTSDGQRSI---SECYLHEPGQ------WAYYHVGDFMVTKPNKPIKIKFS 259
+ + ++S + + +L Q W +G++ V ++ +I+ S
Sbjct: 168 GFESLPLEVSFRSTRTEVYNNRRVFLKSGTQESREDGWLEIELGEYYVGFDDE--EIEMS 225
Query: 260 LAQIDCTHTKGGLCIDSAIIRPTE 283
+ + KGG+ + IRP E
Sbjct: 226 VLETREGGWKGGIIVQGIEIRPKE 249
>UniRef100_Q9C7K0 Hypothetical protein F14G9.14 [Arabidopsis thaliana]
Length = 282
Score = 90.9 bits (224), Expect = 4e-17
Identities = 71/282 (25%), Positives = 127/282 (44%), Gaps = 36/282 (12%)
Query: 31 IPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKFLANKFLGEENLS 90
+PE+CI++IL P D + V+ F A +DFVWE LPS YK L ++ + + +
Sbjct: 4 LPEACIANILAFTSPADAFSSSEVSSVFRLAGDSDFVWEKFLPSDYKSLISQST-DHHWN 62
Query: 91 TMSKKDVYTKLCQPNRFDGGTKEVLLDKYSGQVCLFMSSKSLKITGIDDRRYWIYIPTEE 150
SKK++Y LC D K ++K+SG++ +S++ + IT D YW + +
Sbjct: 63 ISSKKEIYRCLCDSLLIDNARKLFKINKFSGKISYVLSARDISITHSDHASYWSWSNVSD 122
Query: 151 SRFKNVAYLQQMWWVEVIGELE-------------FVFPV--GSYSI-------TFRLQL 188
SRF A L +E+ G+++ + V G+Y + + + +
Sbjct: 123 SRFSESAELIITDRLEIEGKIQTRVLSANTRYGAYLIVKVTKGAYGLDLVPAETSIKSKN 182
Query: 189 GKSSKRLGRRVCNDDQVHG-----WDIKPVRFQLST----SDGQRSISECYLHEPGQWAY 239
G+ SK C D++ + + R ++ DG+R +C W
Sbjct: 183 GQISKSATYLCCLDEKKQQMKRLFYGNREERMAMTVEAVGGDGKRREPKC---RDDGWME 239
Query: 240 YHVGDFMVTKPNKPIKIKFSLAQIDCTHTKGGLCIDSAIIRP 281
+G+F T+ + ++ +L ++ KGG+ ID +RP
Sbjct: 240 IELGEF-ETREGEDDEVNMTLTEVKGYQLKGGILIDGIEVRP 280
>UniRef100_Q9ZVQ8 Putative phloem-specific lectin [Arabidopsis thaliana]
Length = 305
Score = 86.7 bits (213), Expect = 7e-16
Identities = 48/154 (31%), Positives = 82/154 (53%), Gaps = 4/154 (2%)
Query: 19 SNSSSSKTGLGDIPESCISSILMHLDPHDICKLARVNRAFHRASSADFVWESKLPSCYKF 78
S+ S L D+PE C+S I+ P D C LA V++ F A +D VWE +P Y+
Sbjct: 27 SDKSHGVAELDDLPEECVSIIVSFTSPQDACVLASVSKTFASAVKSDIVWEKFIPPEYES 86
Query: 79 LANKFLGEENLSTMSKKDVYTKLCQPN-RFDGGTKEVLLDKYSGQVCLFMSSKSLKITGI 137
L ++ +SKK++Y LC + D G K + ++K + + C+ +S+ +L I
Sbjct: 87 LISQ---SRAFKFLSKKELYFALCDKSVLIDDGKKSLWIEKANAKRCIMISAMNLAIAWG 143
Query: 138 DDRRYWIYIPTEESRFKNVAYLQQMWWVEVIGEL 171
+ + W +IP ++RF+ VA L ++ E+ G +
Sbjct: 144 NSPQSWRWIPDPQARFETVAELLEVCLFEIRGRI 177
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.320 0.135 0.414
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 484,214,320
Number of Sequences: 2790947
Number of extensions: 19295002
Number of successful extensions: 42025
Number of sequences better than 10.0: 102
Number of HSP's better than 10.0 without gapping: 64
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 41908
Number of HSP's gapped (non-prelim): 114
length of query: 291
length of database: 848,049,833
effective HSP length: 126
effective length of query: 165
effective length of database: 496,390,511
effective search space: 81904434315
effective search space used: 81904434315
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 74 (33.1 bits)
Medicago: description of AC149494.4


