

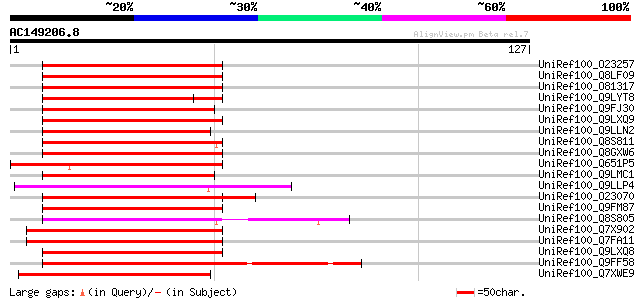
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC149206.8 - phase: 0
(127 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_O23257 Hypothetical protein AT4g13960 [Arabidopsis tha... 61 5e-09
UniRef100_Q8LF09 Hypothetical protein [Arabidopsis thaliana] 58 4e-08
UniRef100_O81317 F6N15.5 protein [Arabidopsis thaliana] 58 4e-08
UniRef100_Q9LYT8 Hypothetical protein F17J16_30 [Arabidopsis tha... 57 7e-08
UniRef100_Q9FJ30 Similarity to heat shock transcription factor H... 57 7e-08
UniRef100_Q9LXQ9 Hypothetical protein T20N10_280 [Arabidopsis th... 57 9e-08
UniRef100_Q9LLN2 Hypothetical protein DUPR11.25 [Oryza sativa] 57 9e-08
UniRef100_Q8S811 Hypothetical protein OSJNBa0020E23.13 [Oryza sa... 57 9e-08
UniRef100_Q8GXW6 Hypothetical protein At3g58930/T20N10_280 [Arab... 57 9e-08
UniRef100_Q651P5 Hypothetical protein OSJNBa0038K02.34 [Oryza sa... 57 9e-08
UniRef100_Q9LMC1 F14D16.22 [Arabidopsis thaliana] 57 1e-07
UniRef100_Q9LLP4 Hypothetical protein DUPR11.8 [Oryza sativa] 57 1e-07
UniRef100_O23070 A_IG005I10.18 protein [Arabidopsis thaliana] 57 1e-07
UniRef100_Q9FM87 Emb|CAB62440.1 [Arabidopsis thaliana] 56 2e-07
UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa] 56 2e-07
UniRef100_Q7X902 P0076O17.1 protein [Oryza sativa] 56 2e-07
UniRef100_Q7FA11 OSJNBa0064D20.2 protein [Oryza sativa] 56 2e-07
UniRef100_Q9LXQ8 Hypothetical protein T20N10_290 [Arabidopsis th... 55 3e-07
UniRef100_Q9FF58 Arabidopsis thaliana genomic DNA, chromosome 5,... 55 3e-07
UniRef100_Q7XWE9 OSJNBa0035O13.11 protein [Oryza sativa] 55 4e-07
>UniRef100_O23257 Hypothetical protein AT4g13960 [Arabidopsis thaliana]
Length = 434
Score = 61.2 bits (147), Expect = 5e-09
Identities = 26/44 (59%), Positives = 35/44 (79%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D +S LPDE++YHILS+L K+ ALTS+LSKRW+ L+ +PN D
Sbjct: 2 DHVSSLPDEVLYHILSFLTTKEAALTSILSKRWRNLFTFVPNLD 45
Score = 38.9 bits (89), Expect = 0.026
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 5/76 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
D +S+LP EVL IL L TK+ +T+ILS RWR + + + D D + +E
Sbjct: 2 DHVSSLPDEVLYHILSFLTTKEAALTSILSKRWRNLFTFV-----PNLDIDDSVFLHPQE 56
Query: 112 EEEEEEEEEEEQQEYV 127
+E+ E ++ ++V
Sbjct: 57 GKEDRYEIQKSFMKFV 72
>UniRef100_Q8LF09 Hypothetical protein [Arabidopsis thaliana]
Length = 441
Score = 58.2 bits (139), Expect = 4e-08
Identities = 23/44 (52%), Positives = 34/44 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DRIS+LPD ++ ILS+LP K + TS+ SK+W+PLW+ +PN +
Sbjct: 4 DRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLE 47
Score = 35.0 bits (79), Expect = 0.38
Identities = 23/60 (38%), Positives = 30/60 (49%), Gaps = 2/60 (3%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHIHYDDDSDSDMEE 109
DRIS LP +L IL LPTK T++ S +WR + + LE + YDD E
Sbjct: 4 DRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLEFDSEDYDDKEQYTFSE 63
>UniRef100_O81317 F6N15.5 protein [Arabidopsis thaliana]
Length = 509
Score = 58.2 bits (139), Expect = 4e-08
Identities = 23/44 (52%), Positives = 34/44 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DRIS+LPD ++ ILS+LP K + TS+ SK+W+PLW+ +PN +
Sbjct: 62 DRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLE 105
Score = 36.2 bits (82), Expect = 0.17
Identities = 24/66 (36%), Positives = 32/66 (48%), Gaps = 2/66 (3%)
Query: 46 RAMPNADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHIHYDDDS 103
R + DRIS LP +L IL LPTK T++ S +WR + + LE + YDD
Sbjct: 56 RVVMGKDRISELPDALLIKILSFLPTKIVVATSVFSKQWRPLWKLVPNLEFDSEDYDDKE 115
Query: 104 DSDMEE 109
E
Sbjct: 116 QYTFSE 121
>UniRef100_Q9LYT8 Hypothetical protein F17J16_30 [Arabidopsis thaliana]
Length = 977
Score = 57.4 bits (137), Expect = 7e-08
Identities = 25/44 (56%), Positives = 33/44 (74%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D +S+LP+E+ HILS+LP K ALTS+LSK W LW+ +PN D
Sbjct: 608 DLVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWKLVPNLD 651
Score = 48.9 bits (115), Expect = 3e-05
Identities = 22/37 (59%), Positives = 30/37 (80%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLW 45
DRIS+LP+EII HI+S+L K+ A S+LSKRW+ L+
Sbjct: 2 DRISNLPNEIICHIVSFLSAKEAAFASILSKRWRNLF 38
Score = 33.1 bits (74), Expect = 1.4
Identities = 18/50 (36%), Positives = 29/50 (58%), Gaps = 3/50 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDD 101
DRIS LP E++ I+ L K+ +ILS RWR + +++ + +DD
Sbjct: 2 DRISNLPNEIICHIVSFLSAKEAAFASILSKRWRNLFTIVIK---LQFDD 48
Score = 32.7 bits (73), Expect = 1.9
Identities = 26/63 (41%), Positives = 34/63 (53%), Gaps = 4/63 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
D +S LP EV IL LPTK +T++LS W L K++ I DDS+ + EEE
Sbjct: 608 DLVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLN-LWKLVPNLDI---DDSEFLLPEEE 663
Query: 112 EEE 114
E
Sbjct: 664 IPE 666
>UniRef100_Q9FJ30 Similarity to heat shock transcription factor HSF30 [Arabidopsis
thaliana]
Length = 540
Score = 57.4 bits (137), Expect = 7e-08
Identities = 26/42 (61%), Positives = 33/42 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DRIS LPD +I HILS+LP K+ A T++L+KRWKPL +PN
Sbjct: 14 DRISGLPDALICHILSFLPTKEAASTTVLAKRWKPLLAFVPN 55
Score = 34.7 bits (78), Expect = 0.50
Identities = 19/52 (36%), Positives = 32/52 (61%), Gaps = 3/52 (5%)
Query: 50 NADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDD 101
+ DRIS LP ++ IL LPTK+ T +L+ RW+ +L + ++++DD
Sbjct: 12 SGDRISGLPDALICHILSFLPTKEAASTTVLAKRWKPLLAFV---PNLNFDD 60
>UniRef100_Q9LXQ9 Hypothetical protein T20N10_280 [Arabidopsis thaliana]
Length = 455
Score = 57.0 bits (136), Expect = 9e-08
Identities = 26/44 (59%), Positives = 33/44 (74%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DR+S+LPD + HILS+LP K IALTS+LSK W LW+ +P D
Sbjct: 2 DRVSNLPDGVRGHILSFLPAKHIALTSVLSKSWLNLWKLIPILD 45
>UniRef100_Q9LLN2 Hypothetical protein DUPR11.25 [Oryza sativa]
Length = 373
Score = 57.0 bits (136), Expect = 9e-08
Identities = 22/41 (53%), Positives = 32/41 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP 49
DRISDLPD++I+H+LS LP + T L++RW+ LWR++P
Sbjct: 6 DRISDLPDDVIHHVLSLLPSRDAVRTCALARRWRDLWRSVP 46
Score = 31.6 bits (70), Expect = 4.2
Identities = 16/35 (45%), Positives = 22/35 (62%)
Query: 51 ADRISALPFEVLSCILYNLPTKQFFVTAILSWRWR 85
ADRIS LP +V+ +L LP++ T L+ RWR
Sbjct: 5 ADRISDLPDDVIHHVLSLLPSRDAVRTCALARRWR 39
>UniRef100_Q8S811 Hypothetical protein OSJNBa0020E23.13 [Oryza sativa]
Length = 949
Score = 57.0 bits (136), Expect = 9e-08
Identities = 26/46 (56%), Positives = 35/46 (75%), Gaps = 2/46 (4%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP--NAD 52
DRIS LPD+I++HI+S+L +Q T +LS+RW+ LWR MP NAD
Sbjct: 90 DRISGLPDKILHHIMSFLNTRQAVQTCVLSRRWRNLWRTMPCINAD 135
Score = 53.5 bits (127), Expect = 1e-06
Identities = 25/46 (54%), Positives = 34/46 (73%), Gaps = 2/46 (4%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP--NAD 52
D ISDLPDEI++ I+S+L +Q T +LS+RW+ LWR +P NAD
Sbjct: 549 DWISDLPDEILHRIMSFLNARQAVQTCVLSRRWRNLWRTVPCINAD 594
Score = 33.9 bits (76), Expect = 0.85
Identities = 17/36 (47%), Positives = 23/36 (63%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRV 87
DRIS LP ++L I+ L T+Q T +LS RWR +
Sbjct: 90 DRISGLPDKILHHIMSFLNTRQAVQTCVLSRRWRNL 125
Score = 31.2 bits (69), Expect = 5.5
Identities = 23/84 (27%), Positives = 34/84 (40%), Gaps = 15/84 (17%)
Query: 18 IIYHILSYLPPKQIALTSLLSKRWKPLWRAM--------------PNADRISALPFEVLS 63
+I+H+ +P + W P WR + D IS LP E+L
Sbjct: 502 VIFHMAIKVPHAHHLFDETPLRAWPP-WRGVLAVARRKVKTPAHAAGMDWISDLPDEILH 560
Query: 64 CILYNLPTKQFFVTAILSWRWRRV 87
I+ L +Q T +LS RWR +
Sbjct: 561 RIMSFLNARQAVQTCVLSRRWRNL 584
>UniRef100_Q8GXW6 Hypothetical protein At3g58930/T20N10_280 [Arabidopsis thaliana]
Length = 482
Score = 57.0 bits (136), Expect = 9e-08
Identities = 26/44 (59%), Positives = 33/44 (74%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DR+S+LPD + HILS+LP K IALTS+LSK W LW+ +P D
Sbjct: 2 DRVSNLPDGVRGHILSFLPAKHIALTSVLSKSWLNLWKLIPILD 45
>UniRef100_Q651P5 Hypothetical protein OSJNBa0038K02.34 [Oryza sativa]
Length = 591
Score = 57.0 bits (136), Expect = 9e-08
Identities = 27/63 (42%), Positives = 39/63 (61%), Gaps = 11/63 (17%)
Query: 1 MAAPPTTTDRISD-----------LPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP 49
MA+PPT+ I+ LPD++++H+LS+LP A TSLLS+RW+ LW +MP
Sbjct: 1 MASPPTSESAITSAGSASASGFDCLPDDLVHHVLSFLPAPDAACTSLLSRRWRNLWVSMP 60
Query: 50 NAD 52
D
Sbjct: 61 CLD 63
>UniRef100_Q9LMC1 F14D16.22 [Arabidopsis thaliana]
Length = 269
Score = 56.6 bits (135), Expect = 1e-07
Identities = 27/42 (64%), Positives = 33/42 (78%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPN 50
DRI+ L DEII HILS+LP K+ ALTS+LSKRW+ L+ PN
Sbjct: 2 DRINGLSDEIICHILSFLPTKESALTSVLSKRWRNLFALTPN 43
Score = 38.5 bits (88), Expect = 0.034
Identities = 22/51 (43%), Positives = 31/51 (60%), Gaps = 3/51 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDD 102
DRI+ L E++ IL LPTK+ +T++LS RWR N T ++H DD
Sbjct: 2 DRINGLSDEIICHILSFLPTKESALTSVLSKRWR---NLFALTPNLHLVDD 49
>UniRef100_Q9LLP4 Hypothetical protein DUPR11.8 [Oryza sativa]
Length = 476
Score = 56.6 bits (135), Expect = 1e-07
Identities = 27/73 (36%), Positives = 42/73 (56%), Gaps = 5/73 (6%)
Query: 2 AAPPTTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRA-----MPNADRISA 56
A PPT DRI LPD ++ HIL +LPP + T +L++RW+ LW++ + N D
Sbjct: 85 APPPTGGDRIGGLPDGVLQHILGFLPPHEAVRTCVLARRWRHLWKSVAALRITNWDWRKV 144
Query: 57 LPFEVLSCILYNL 69
+P E +++L
Sbjct: 145 VPMEEFRYFVHHL 157
Score = 32.0 bits (71), Expect = 3.2
Identities = 20/58 (34%), Positives = 27/58 (46%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEE 109
DRI LP VL IL LP + T +L+ RWR + + ++D MEE
Sbjct: 92 DRIGGLPDGVLQHILGFLPPHEAVRTCVLARRWRHLWKSVAALRITNWDWRKVVPMEE 149
>UniRef100_O23070 A_IG005I10.18 protein [Arabidopsis thaliana]
Length = 773
Score = 56.6 bits (135), Expect = 1e-07
Identities = 28/52 (53%), Positives = 35/52 (66%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNADRISALPFE 60
D IS LPD +I HILS+LP K A T++L+KRWKPL MPN D + F+
Sbjct: 279 DGISGLPDAMICHILSFLPTKVAASTTVLAKRWKPLLAFMPNLDFDESFRFD 330
Score = 51.6 bits (122), Expect = 4e-06
Identities = 22/44 (50%), Positives = 29/44 (65%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
D+ S LPDE++ +LS+LP K TSLLS RWK LW +P +
Sbjct: 2 DKTSQLPDELLVKVLSFLPTKDAVRTSLLSMRWKSLWMWLPKLE 45
Score = 33.1 bits (74), Expect = 1.4
Identities = 20/50 (40%), Positives = 29/50 (58%), Gaps = 2/50 (4%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHIHY 99
D+ S LP E+L +L LPTK T++LS RW+ + + LE + HY
Sbjct: 2 DKTSQLPDELLVKVLSFLPTKDAVRTSLLSMRWKSLWMWLPKLEYDFRHY 51
>UniRef100_Q9FM87 Emb|CAB62440.1 [Arabidopsis thaliana]
Length = 430
Score = 56.2 bits (134), Expect = 2e-07
Identities = 23/44 (52%), Positives = 34/44 (77%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DRIS LPD++++ ILS++P K + T+LLSKRW+ LW+ +P D
Sbjct: 2 DRISLLPDDVVFKILSFVPTKVVVSTNLLSKRWRYLWKHVPKLD 45
Score = 33.9 bits (76), Expect = 0.85
Identities = 23/57 (40%), Positives = 31/57 (54%), Gaps = 3/57 (5%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDME 108
DRIS LP +V+ IL +PTK T +LS RWR + + + + Y D S D E
Sbjct: 2 DRISLLPDDVVFKILSFVPTKVVVSTNLLSKRWRYLWKHVPK---LDYRDPSLVDTE 55
>UniRef100_Q8S805 Putative copia-type polyprotein [Oryza sativa]
Length = 1803
Score = 55.8 bits (133), Expect = 2e-07
Identities = 33/79 (41%), Positives = 45/79 (56%), Gaps = 10/79 (12%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP--NADRISALPFEVLSCIL 66
DR+SDLPDEI++ I+S+L +Q T +LS+RW+ LW +P NAD F I
Sbjct: 1424 DRLSDLPDEILHRIMSFLNARQAVQTCVLSRRWRNLWHTVPCINAD------FVEFDSIG 1477
Query: 67 YNLPTKQF--FVTAILSWR 83
Y P F FV +L +R
Sbjct: 1478 YQGPVVPFKRFVNRLLEFR 1496
Score = 33.5 bits (75), Expect = 1.1
Identities = 19/63 (30%), Positives = 32/63 (50%), Gaps = 2/63 (3%)
Query: 40 RWKPLWRAMPNADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKI--LETNHI 97
R +P DR+S LP E+L I+ L +Q T +LS RWR + + + + + +
Sbjct: 1412 RARPPMEDAAGRDRLSDLPDEILHRIMSFLNARQAVQTCVLSRRWRNLWHTVPCINADFV 1471
Query: 98 HYD 100
+D
Sbjct: 1472 EFD 1474
>UniRef100_Q7X902 P0076O17.1 protein [Oryza sativa]
Length = 384
Score = 55.8 bits (133), Expect = 2e-07
Identities = 25/48 (52%), Positives = 35/48 (72%)
Query: 5 PTTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
P TDR+S LPD +++ ILS L +Q+ TS+LSKRW+ LWR++P D
Sbjct: 31 PPATDRLSALPDALLHTILSSLKGRQMVQTSVLSKRWRHLWRSVPCLD 78
Score = 38.9 bits (89), Expect = 0.026
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 6/62 (9%)
Query: 36 LLSKRWKPLWRAM------PNADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLN 89
L+ W+P R P DR+SALP +L IL +L +Q T++LS RWR +
Sbjct: 13 LILHPWRPRGRGAVQTPSPPATDRLSALPDALLHTILSSLKGRQMVQTSVLSKRWRHLWR 72
Query: 90 KI 91
+
Sbjct: 73 SV 74
>UniRef100_Q7FA11 OSJNBa0064D20.2 protein [Oryza sativa]
Length = 388
Score = 55.8 bits (133), Expect = 2e-07
Identities = 25/48 (52%), Positives = 35/48 (72%)
Query: 5 PTTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
P TDR+S LPD +++ ILS L +Q+ TS+LSKRW+ LWR++P D
Sbjct: 35 PPATDRLSALPDALLHTILSSLKGRQMVQTSVLSKRWRHLWRSVPCLD 82
Score = 38.9 bits (89), Expect = 0.026
Identities = 22/62 (35%), Positives = 32/62 (51%), Gaps = 6/62 (9%)
Query: 36 LLSKRWKPLWRAM------PNADRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLN 89
L+ W+P R P DR+SALP +L IL +L +Q T++LS RWR +
Sbjct: 17 LILHPWRPRGRGAVQTPSPPATDRLSALPDALLHTILSSLKGRQMVQTSVLSKRWRHLWR 76
Query: 90 KI 91
+
Sbjct: 77 SV 78
>UniRef100_Q9LXQ8 Hypothetical protein T20N10_290 [Arabidopsis thaliana]
Length = 618
Score = 55.5 bits (132), Expect = 3e-07
Identities = 25/44 (56%), Positives = 32/44 (71%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNAD 52
DR+S+LP+E+ HILS+LP K ALTS+LSK W LW+ N D
Sbjct: 2 DRVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWKFETNLD 45
Score = 39.3 bits (90), Expect = 0.020
Identities = 28/76 (36%), Positives = 38/76 (49%), Gaps = 5/76 (6%)
Query: 52 DRISALPFEVLSCILYNLPTKQFFVTAILSWRWRRVLNKILETNHIHYDDDSDSDMEEEE 111
DR+S LP EV IL LPTK +T++LS W + ETN D D + EE
Sbjct: 2 DRVSNLPEEVRCHILSFLPTKHAALTSVLSKSWLNLWK--FETN---LDIDDSDFLHPEE 56
Query: 112 EEEEEEEEEEEQQEYV 127
+ E +E + E+V
Sbjct: 57 GKAERDEIRQSFVEFV 72
>UniRef100_Q9FF58 Arabidopsis thaliana genomic DNA, chromosome 5, P1 clone:MUP24
[Arabidopsis thaliana]
Length = 388
Score = 55.5 bits (132), Expect = 3e-07
Identities = 32/78 (41%), Positives = 47/78 (60%), Gaps = 2/78 (2%)
Query: 9 DRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMPNADRISALPFEVLSCILYN 68
DRIS LPDE++ I+S++P K TS+LSKRW+ LW+ +P + P + IL N
Sbjct: 2 DRISGLPDELLVKIISFVPTKVAVSTSILSKRWESLWKWVPKLECDCTEP-ALRDFILKN 60
Query: 69 LPTKQFFVTAILSWRWRR 86
LP + + + L R+RR
Sbjct: 61 LPLQARIIES-LYLRFRR 77
>UniRef100_Q7XWE9 OSJNBa0035O13.11 protein [Oryza sativa]
Length = 506
Score = 55.1 bits (131), Expect = 4e-07
Identities = 24/47 (51%), Positives = 32/47 (68%)
Query: 3 APPTTTDRISDLPDEIIYHILSYLPPKQIALTSLLSKRWKPLWRAMP 49
AP DRIS LPD ++ ++S LP K A T+ LS+RW+PLWR+ P
Sbjct: 45 APHDAVDRISALPDALLRRVVSRLPVKDAARTAALSRRWRPLWRSTP 91
Score = 35.4 bits (80), Expect = 0.29
Identities = 29/81 (35%), Positives = 39/81 (47%), Gaps = 8/81 (9%)
Query: 10 RISDLPDEI---IYHILSYLPPKQIALTSLLSKRWKPLWRAMPN--ADRISALPFEVLSC 64
R D PD++ + +LSY+ AL L+ P+ DRISALP +L
Sbjct: 7 RRGDDPDDVQGTVARVLSYI---HYALPDPPVSAAARLYALAPHDAVDRISALPDALLRR 63
Query: 65 ILYNLPTKQFFVTAILSWRWR 85
++ LP K TA LS RWR
Sbjct: 64 VVSRLPVKDAARTAALSRRWR 84
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.315 0.133 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 239,657,283
Number of Sequences: 2790947
Number of extensions: 10356459
Number of successful extensions: 473542
Number of sequences better than 10.0: 6571
Number of HSP's better than 10.0 without gapping: 6207
Number of HSP's successfully gapped in prelim test: 402
Number of HSP's that attempted gapping in prelim test: 298339
Number of HSP's gapped (non-prelim): 74184
length of query: 127
length of database: 848,049,833
effective HSP length: 103
effective length of query: 24
effective length of database: 560,582,292
effective search space: 13453975008
effective search space used: 13453975008
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 67 (30.4 bits)
Medicago: description of AC149206.8


