

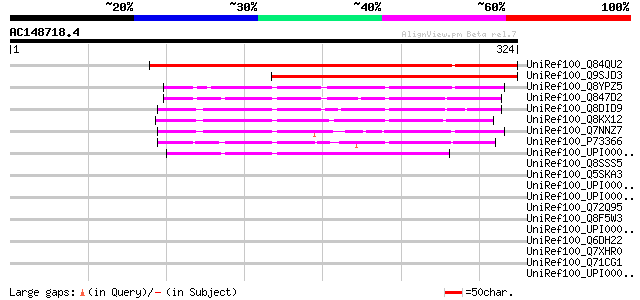
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148718.4 + phase: 0
(324 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q84QU2 Hypothetical protein P0410E11.116 [Oryza sativa] 344 2e-93
UniRef100_Q9SJD3 Hypothetical protein At2g04360 [Arabidopsis tha... 227 3e-58
UniRef100_Q8YPZ5 Alr4045 protein [Anabaena sp.] 130 6e-29
UniRef100_Q847D2 Hypothetical protein [Nodularia spumigena] 124 5e-27
UniRef100_Q8DID9 Tll1651 protein [Synechococcus elongatus] 117 3e-25
UniRef100_Q8KX12 Hypothetical protein slr1215 [Synechococcus sp.] 113 8e-24
UniRef100_Q7NNZ7 Gll0261 protein [Gloeobacter violaceus] 99 2e-19
UniRef100_P73366 Slr1215 protein [Synechocystis sp.] 94 5e-18
UniRef100_UPI000031FB35 UPI000031FB35 UniRef100 entry 59 2e-07
UniRef100_Q8SSS5 Similar to Staphylococcus epidermidis ATCC 1222... 37 0.95
UniRef100_Q5SKA3 Hypothetical protein TTHA0745 [Thermus thermoph... 36 1.6
UniRef100_UPI00002D5041 UPI00002D5041 UniRef100 entry 35 2.1
UniRef100_UPI00003144F1 UPI00003144F1 UniRef100 entry 35 2.8
UniRef100_Q72Q95 Putative lipoprotein [Leptospira interrogans] 35 3.6
UniRef100_Q8F5W3 Putative lipoprotein [Leptospira interrogans] 34 4.7
UniRef100_UPI0000342270 UPI0000342270 UniRef100 entry 34 6.1
UniRef100_Q6DH22 Hypothetical protein [Brachydanio rerio] 34 6.1
UniRef100_Q7XHR0 Putative FKBP-type peptidyl-prolyl cis-trans is... 34 6.1
UniRef100_Q71CG1 Class V chitin synthase [Coccidioides posadasii] 34 6.1
UniRef100_UPI00001A135F UPI00001A135F UniRef100 entry 33 8.0
>UniRef100_Q84QU2 Hypothetical protein P0410E11.116 [Oryza sativa]
Length = 357
Score = 344 bits (883), Expect = 2e-93
Identities = 156/235 (66%), Positives = 190/235 (80%), Gaps = 1/235 (0%)
Query: 90 DWTTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIM 149
DWTTS+L+F +WA L+YY+ L+PNQTP RD YFL+KL NLKGDDGFRMN+VLV WYIM
Sbjct: 122 DWTTSVLIFGIWAGLMYYIFQLAPNQTPYRDTYFLQKLCNLKGDDGFRMNDVLVPLWYIM 181
Query: 150 GFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKT 209
G WPLVYSMLLLPTGRSSKS +PVWPFL S GG YAL+PYFVLWKPPPPP++E E+
Sbjct: 182 GLWPLVYSMLLLPTGRSSKSKIPVWPFLILSCIGGAYALIPYFVLWKPPPPPIDEEEIGQ 241
Query: 210 WPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTL 269
WPL FLESK+TA + A G+G+I+YA AG + W+EF +Y RESK IHIT +DF LLS
Sbjct: 242 WPLKFLESKLTAGVTFAVGLGLIVYAAKAGGEDWQEFIRYFRESKLIHITCLDFCLLSAF 301
Query: 270 APFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAVAQSPAESE 324
+PFWVYND+TAR+W GSWLLP++LIP +GP+LY+LLRPSLS+ A P++ +
Sbjct: 302 SPFWVYNDLTARRW-KNGSWLLPLALIPFVGPSLYLLLRPSLSSLLAATGPSDDK 355
>UniRef100_Q9SJD3 Hypothetical protein At2g04360 [Arabidopsis thaliana]
Length = 295
Score = 227 bits (579), Expect = 3e-58
Identities = 100/157 (63%), Positives = 122/157 (77%)
Query: 168 KSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESKITAMILLAS 227
+S P WPF+ SFFGG+YALLPYF LW PP PPV E EL+ WPLN LESK+TA + L +
Sbjct: 136 ESKTPAWPFVVLSFFGGVYALLPYFALWNPPSPPVSETELRQWPLNVLESKVTAGVTLVA 195
Query: 228 GIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDMTARKWFDKG 287
G+GII+Y+ + W EF+QY RESKFIH+TS+DF LLS APFWVYNDMT RKWFDKG
Sbjct: 196 GLGIILYSVVGNAGDWTEFYQYFRESKFIHVTSLDFCLLSAFAPFWVYNDMTTRKWFDKG 255
Query: 288 SWLLPVSLIPLLGPALYILLRPSLSTSAVAQSPAESE 324
SWLLPVS+IP LGP+LY+LLRP++S + + A S+
Sbjct: 256 SWLLPVSVIPFLGPSLYLLLRPAVSETIAPKDTASSD 292
>UniRef100_Q8YPZ5 Alr4045 protein [Anabaena sp.]
Length = 226
Score = 130 bits (326), Expect = 6e-29
Identities = 80/218 (36%), Positives = 116/218 (52%), Gaps = 12/218 (5%)
Query: 99 LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSM 158
+LW +Y L+P P D + L K NL +N ++++ + +MG WP++YS
Sbjct: 9 VLWLGFTFYAFVLAPPDQP--DTFELIK--NLSTGQWQGVNPLIIALFNLMGIWPMIYSA 64
Query: 159 LLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESK 218
L+ GR K +P W F +ASF GI+ALLPY L +P P + K + L+S+
Sbjct: 65 LMFIDGRGQK--IPAWLFATASFGVGIFALLPYLTLREPNP---QFPGTKNTLIKLLDSR 119
Query: 219 ITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDM 278
+T MIL ++ Y G+ W F Q + +FIH+ S+DF LLS L P + +DM
Sbjct: 120 VTGMILTIVAAILVAYGLQQGD--WSNFLQQWQTKRFIHVMSLDFCLLSLLFPALLGDDM 177
Query: 279 TARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTSAV 316
T R W K +LIPL GP LY+ LRP L ++V
Sbjct: 178 TRRGW-QKPQLFWVFALIPLFGPLLYLCLRPPLPEASV 214
>UniRef100_Q847D2 Hypothetical protein [Nodularia spumigena]
Length = 219
Score = 124 bits (310), Expect = 5e-27
Identities = 80/217 (36%), Positives = 124/217 (56%), Gaps = 14/217 (6%)
Query: 99 LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSM 158
LLW + Y +P P D + L K L++ +G +N ++++ + +MG WP++YS
Sbjct: 9 LLWLGFLTYAFLFAPPDQP--DTFELIKNLSVGKWEG--INPLVIALFNLMGIWPVIYSA 64
Query: 159 LLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESK 218
+L GR K +P WPF +ASF G +ALLPY VL +P E + K+ L L+S+
Sbjct: 65 VLFMDGRGQK--IPAWPFATASFAVGAFALLPYLVLREPNQ---EFSGKKSLWLKLLDSR 119
Query: 219 ITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDM 278
+T +I L G I++ GL G+ W F Q + S+FIH+ S+DF +LS L P + +DM
Sbjct: 120 VTGLI-LTIGAAILVRYGLQGD--WGNFVQQWQTSRFIHVMSLDFCVLSLLFPALLGDDM 176
Query: 279 TARKWFDKGSWLLPVSLIPLLGPALYILLR-PSLSTS 314
AR+ + ++LIPL GP +Y+ +R P L T+
Sbjct: 177 -ARRGMKNQVFFWLIALIPLFGPLIYLSVRSPLLETN 212
>UniRef100_Q8DID9 Tll1651 protein [Synechococcus elongatus]
Length = 232
Score = 117 bits (294), Expect = 3e-25
Identities = 77/220 (35%), Positives = 118/220 (53%), Gaps = 13/220 (5%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
+ L +LW LI Y +P P L + L + +N ++VS + +MG WPL
Sbjct: 21 LFLAVLWVGLIVYSFGFAPPDQPQT----LTLIQRLATGNWQGINPLIVSLFNLMGVWPL 76
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
+Y+ LL+ G+ + P WPF + SF G +ALLPY +L +P P + AEL +
Sbjct: 77 IYTALLVVDGQGQR--FPAWPFAAFSFVVGAFALLPYLILRQPAP--LFTAELNRG-VRL 131
Query: 215 LESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWV 274
ES+IT + + I ++ Y AG+ W++F+Q S+FIH+ ++DF LLS L P +
Sbjct: 132 WESRITGIGIFLLAIVLLSYGLFAGD--WQDFWQQWHSSRFIHVMTLDFCLLSLLLPVLL 189
Query: 275 YNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPSLSTS 314
+D R+ +K W SL+P +G Y+LLRP L S
Sbjct: 190 VDDW-QRRGLEKSPWRW-WSLVPFVGALGYLLLRPPLILS 227
>UniRef100_Q8KX12 Hypothetical protein slr1215 [Synechococcus sp.]
Length = 231
Score = 113 bits (282), Expect = 8e-24
Identities = 73/217 (33%), Positives = 115/217 (52%), Gaps = 13/217 (5%)
Query: 94 SILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWP 153
SI L LW A + Y +P P L+ + L G +N ++++ + IMG +P
Sbjct: 4 SIFLGSLWLAFVLYAFLGAPPNQPET----LELITALSGGQWDGINPLIIALFNIMGIFP 59
Query: 154 LVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLN 213
++Y LL G K +P W F + SFF G +ALLPY L + P + K W L
Sbjct: 60 VMYGALLFLDGDRQK--LPAWVFAAGSFFLGAFALLPYLALRRDNPTFTGQ---KNWWLR 114
Query: 214 FLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFW 273
+ +S++ + L + +G++IY AG+ W +F +FIH+ S+DF LL+ L P
Sbjct: 115 WQDSRLLGLFCLVALLGLLIYGFSAGD--WGDFGTRFWGDRFIHVMSLDFCLLALLFPTL 172
Query: 274 VYNDMTARKWFDKGSWLL-PVSLIPLLGPALYILLRP 309
+ +D +AR+ + +WLL ++ P LG ALY+ LRP
Sbjct: 173 IKDD-SARRGLGEQNWLLWAIAFTPPLGAALYLALRP 208
>UniRef100_Q7NNZ7 Gll0261 protein [Gloeobacter violaceus]
Length = 219
Score = 98.6 bits (244), Expect = 2e-19
Identities = 73/229 (31%), Positives = 112/229 (48%), Gaps = 24/229 (10%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
I L LW + Y P +T S L++ +L R + V+ + +MG PL
Sbjct: 8 IALAALWLGFVAYAFLGGPPETRSP----LQQAASLADLALGRGEPLSVALFNLMGLIPL 63
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFV------LWKPPPPPVEEAELK 208
Y LL+P K +P WPF ASF G +ALLPY + +W PP A L
Sbjct: 64 AYLFLLVPDAHGEK--LPAWPFAVASFAVGAFALLPYLICRRPHGVWPHIPPAGRAARL- 120
Query: 209 TWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLST 268
+S+ A+ ++A G+ + + GL + W F + R S+F+H+ +DF + +
Sbjct: 121 ------FDSRWFALAVMA-GVAGLFWFGLT-QGNWAAFAEQWRTSRFVHVMGLDFLVSTA 172
Query: 269 LAPFWVYNDMTARKWFDKGSWLLPVS-LIPLLGPALYILLRPSLSTSAV 316
L P + +D+ R K +W L ++ +PL GP LY++LRPSL AV
Sbjct: 173 LLPLLIGDDLRRRS--VKRTWPLWIACALPLFGPLLYLVLRPSLPARAV 219
>UniRef100_P73366 Slr1215 protein [Synechocystis sp.]
Length = 222
Score = 94.0 bits (232), Expect = 5e-18
Identities = 66/224 (29%), Positives = 108/224 (47%), Gaps = 23/224 (10%)
Query: 95 ILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPL 154
+ L LW AL+ + +P P + + +K L + D +N V+++ + +MG WP+
Sbjct: 6 LALACLWLALVIFAFGFAPPSNP-QTLVLIKSLSLGQWQD---INPVIIALFNLMGIWPM 61
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
Y+ +LL + K P WPF++ SFF G +ALLPY V W PP P + A P N
Sbjct: 62 TYAAMLLGDRQGRK--FPAWPFVAGSFFLGAFALLPYLVFW-PPTAPTDPA-----PSNL 113
Query: 215 LESKIT--------AMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLL 266
SK+T +L I + A G+ W ++ Q + ++FI + S DF L
Sbjct: 114 GSSKLTKFWRSPWLGRVLFLLAIACVSGAVFRGD--WADYGQQLQTNQFIAVMSSDFLCL 171
Query: 267 STLAPFWVYNDMTARKWFDKGSWLLPVSLIPLLGPALYILLRPS 310
+ P + ++ +K L+ S +PL G Y+ +RP+
Sbjct: 172 TLAFPLVLAQELKHQK-ASPSLLLVLGSTVPLFGALAYLSIRPN 214
>UniRef100_UPI000031FB35 UPI000031FB35 UniRef100 entry
Length = 204
Score = 58.5 bits (140), Expect = 2e-07
Identities = 48/183 (26%), Positives = 79/183 (42%), Gaps = 6/183 (3%)
Query: 101 WAALIYYVSFLSPNQTPSRDMYFLKKLLNLKGDDGFRMNEVLVSEWYIMGFWPLVYSMLL 160
W +L + +F +P T + D+ +K ++ N + + + +G P Y+ LL
Sbjct: 24 WVSLGSFATFGAPAGTSAFDLELVKTIVGAPFSGA--ANPLFEALFNALGVVPATYACLL 81
Query: 161 LPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNFLESKIT 220
+P + K +P + ASF G +AL PY VL +P P + +EL N ESKI
Sbjct: 82 MPGAKDQK--IPAALCVGASFALGFFALGPYLVLREPRTAPTKRSELGFVTRNVWESKIN 139
Query: 221 AMILLASGIGIIIYA-GLAGEDVWKEFFQYCRESKFIH-ITSIDFTLLSTLAPFWVYNDM 278
A+ L + Y D F +E + +++ D +LS + DM
Sbjct: 140 AVFLAGFAAYLAFYGFSHLNADTLAAFASLFKEQSMLACVSTCDLVVLSLCVGTAIAEDM 199
Query: 279 TAR 281
T R
Sbjct: 200 TRR 202
>UniRef100_Q8SSS5 Similar to Staphylococcus epidermidis ATCC 12228. Streptococcal
hemagglutinin protein [Dictyostelium discoideum]
Length = 1297
Score = 36.6 bits (83), Expect = 0.95
Identities = 18/74 (24%), Positives = 35/74 (46%)
Query: 21 KKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFRRKSSSSSIVICNDSNKQNSS 80
K+ Q +++++ CRL+ F PL + F F+ K+S + I N++N N++
Sbjct: 378 KRKTQKDVSLISIMNWCRLNGFTPLTTHQESFYNLIDPFKNKTSDNFITTTNNNNNNNNN 437
Query: 81 VEEKEEVKRDWTTS 94
+ TT+
Sbjct: 438 NNNTDTTTTTTTTT 451
>UniRef100_Q5SKA3 Hypothetical protein TTHA0745 [Thermus thermophilus HB8]
Length = 518
Score = 35.8 bits (81), Expect = 1.6
Identities = 23/61 (37%), Positives = 27/61 (43%), Gaps = 3/61 (4%)
Query: 155 VYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWKPPPPPVEEAELKTWPLNF 214
+Y + LLP R S P P A FF L VL+ P PPP E A+ W F
Sbjct: 179 LYRLRLLPESRLHLSGRPTPPKTLAEFFP---PYLERVVLYLPDPPPPEAAQATLWLAGF 235
Query: 215 L 215
L
Sbjct: 236 L 236
>UniRef100_UPI00002D5041 UPI00002D5041 UniRef100 entry
Length = 429
Score = 35.4 bits (80), Expect = 2.1
Identities = 30/118 (25%), Positives = 52/118 (43%), Gaps = 18/118 (15%)
Query: 91 WTTSILLF--LLWAALIYYVSFLSPNQTPSRDMYFLKKLLNLK------GDDGFRMNEVL 142
W ++LLF + W A+ +F + Q + ++F+ L + + F N VL
Sbjct: 174 WLIALLLFFPIFWMAI---TAFKTEQQAYASSLFFIPTLDSFREVFARSNYFAFAWNSVL 230
Query: 143 VSEWY----IMGFWPLVYSMLLLPTGRSSKSSVPVWPFLSASFFGGIYALLPYFVLWK 196
+S ++ P Y+M P R+ K V +W LS + L+P ++LWK
Sbjct: 231 ISAGVTVISLLFAVPAAYAMAFFPNHRTQK--VLLW-MLSTKMMPSVGVLVPIYLLWK 285
>UniRef100_UPI00003144F1 UPI00003144F1 UniRef100 entry
Length = 244
Score = 35.0 bits (79), Expect = 2.8
Identities = 21/84 (25%), Positives = 41/84 (48%), Gaps = 8/84 (9%)
Query: 1 MVLVVAKPNLISCNFPSLKTKKSIQPHITVVASGCCCRLSSFKPLVAMKTPFVAATTSFR 60
+ + +AK ++SCN PS KTK + T CR++ + + P+++ + R
Sbjct: 67 VAIALAKTRIMSCNLPSYKTKNDNNGYTTQE-----CRINPRLGFIYKQVPYISLRSIAR 121
Query: 61 RKSSSSSIVICNDSNKQNSSVEEK 84
K VI ++ NK+ ++ +K
Sbjct: 122 NKEID---VIWDNYNKKLETIRQK 142
>UniRef100_Q72Q95 Putative lipoprotein [Leptospira interrogans]
Length = 673
Score = 34.7 bits (78), Expect = 3.6
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 11/53 (20%)
Query: 266 LSTLAPFWVY-----------NDMTARKWFDKGSWLLPVSLIPLLGPALYILL 307
L L PF +Y D T +W K W++ + L+P+L PA+Y+++
Sbjct: 579 LGFLLPFAIYICWTSMSLLDLKDRTDLEWIRKIYWIVTIILVPILSPAIYLII 631
>UniRef100_Q8F5W3 Putative lipoprotein [Leptospira interrogans]
Length = 673
Score = 34.3 bits (77), Expect = 4.7
Identities = 16/53 (30%), Positives = 27/53 (50%), Gaps = 11/53 (20%)
Query: 266 LSTLAPFWVY-----------NDMTARKWFDKGSWLLPVSLIPLLGPALYILL 307
L L PF +Y D T +W K W++ + L+P+L PA+Y+++
Sbjct: 579 LGFLLPFAIYICWTSMSLLDLKDRTDLEWIRKIYWIVTIILVPILSPAIYLIV 631
>UniRef100_UPI0000342270 UPI0000342270 UniRef100 entry
Length = 415
Score = 33.9 bits (76), Expect = 6.1
Identities = 20/79 (25%), Positives = 40/79 (50%), Gaps = 6/79 (7%)
Query: 221 AMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFTLLSTLAPFWVYNDMTA 280
A+ +LA+ +G + Y + W++F+ +FI + + L S L F V + T+
Sbjct: 13 ALGILATILGFVFYTSSSSHPFWQKFY------RFIPALLLCYFLPSLLNTFGVIDGHTS 66
Query: 281 RKWFDKGSWLLPVSLIPLL 299
+ ++ +LLP L+ L+
Sbjct: 67 QLYYVASRYLLPACLVLLI 85
>UniRef100_Q6DH22 Hypothetical protein [Brachydanio rerio]
Length = 306
Score = 33.9 bits (76), Expect = 6.1
Identities = 22/78 (28%), Positives = 33/78 (42%), Gaps = 4/78 (5%)
Query: 207 LKTWPLNFLESKITAMILLASGIGIIIYAGLAGEDVWKEFFQYCRESKFIHITSIDFT-L 265
L+ WP ++ A + S + I + G W + QYC + K TSI + L
Sbjct: 92 LRHWPFGSEGCQLHAFQGMVSILAAISFLGAVA---WDRYHQYCTKQKMFWSTSITISCL 148
Query: 266 LSTLAPFWVYNDMTARKW 283
+ LA FW + A W
Sbjct: 149 IWILAVFWAAMPLPAIGW 166
>UniRef100_Q7XHR0 Putative FKBP-type peptidyl-prolyl cis-trans isomerase; protein
[Oryza sativa]
Length = 258
Score = 33.9 bits (76), Expect = 6.1
Identities = 40/139 (28%), Positives = 59/139 (41%), Gaps = 22/139 (15%)
Query: 51 PFVAATTSFRRKSSSSSIVICNDSNKQNSSVEEKEEVKRDWTTSILLFLLWAALIYYVSF 110
PF A+ S RRK +S + + SV E +R L LL A+ V+F
Sbjct: 26 PFRTASASSRRKGTSGFVCALGCDGEGKKSVAEGTVRRR-----AALALLLASPAMSVAF 80
Query: 111 LSPNQTPSRDMYFLKKLL---------NLKGDD-------GFRMNEVLVSEWYIMGFWPL 154
+ +T SR+ Y ++LL N DD GF + +V+ S+ YI L
Sbjct: 81 SAHGKTKSRNPYDERRLLQQNKKIQEANRAPDDFPNFIREGFEV-KVVTSDNYITRDSGL 139
Query: 155 VYSMLLLPTGRSSKSSVPV 173
+Y + + TG S K V
Sbjct: 140 LYEDIKVGTGNSPKDGQQV 158
>UniRef100_Q71CG1 Class V chitin synthase [Coccidioides posadasii]
Length = 1772
Score = 33.9 bits (76), Expect = 6.1
Identities = 22/86 (25%), Positives = 47/86 (54%), Gaps = 6/86 (6%)
Query: 83 EKEEVKRDW----TTSILLFLLWAALIYYVSFLSPNQTPSRDMYFLKKLLNL-KGDDGFR 137
++ +V+ W T +L+FLL A++++Y+ FL P++D + K ++ +G++ F
Sbjct: 767 KRPDVRMAWREKVTLILLIFLLNASIVFYIVFLGDLLCPNKDKVWTDKEVSYHQGENDFY 826
Query: 138 MNEVLVSEWYIMGFWPLVYSMLLLPT 163
++ V + I FW + ++ PT
Sbjct: 827 VS-VHGKVYDISKFWKITHTTNRYPT 851
>UniRef100_UPI00001A135F UPI00001A135F UniRef100 entry
Length = 365
Score = 33.5 bits (75), Expect = 8.0
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 7/59 (11%)
Query: 221 AMILLASGIGIIIYAGLAGEDVWKEFFQYCRESK------FIHITSIDFTLLSTLAPFW 273
A I L + G+ + GLAG + + YC++ K +H+ D LL TL PFW
Sbjct: 39 ARIFLPAVFGLSLVIGLAGNALIVAVYAYCKQLKTMTDTFILHLAVADLLLLLTL-PFW 96
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.137 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,386,615
Number of Sequences: 2790947
Number of extensions: 22949563
Number of successful extensions: 70450
Number of sequences better than 10.0: 24
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 70419
Number of HSP's gapped (non-prelim): 25
length of query: 324
length of database: 848,049,833
effective HSP length: 127
effective length of query: 197
effective length of database: 493,599,564
effective search space: 97239114108
effective search space used: 97239114108
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 75 (33.5 bits)
Medicago: description of AC148718.4


