

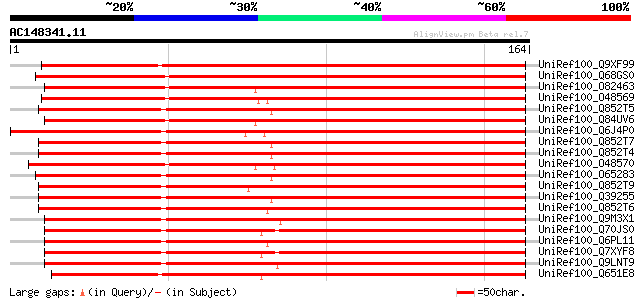
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC148341.11 - phase: 0
(164 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9XF99 Skp1 [Medicago sativa] 253 1e-66
UniRef100_Q68GS0 SKP1 [Nicotiana tabacum] 239 1e-62
UniRef100_O82463 SKP1-like protein [Nicotiana clevelandii] 228 3e-59
UniRef100_O48569 Fimbriata-associated protein [Antirrhinum majus] 228 6e-59
UniRef100_Q852T5 Kinetochore protein [Brassica juncea] 226 1e-58
UniRef100_Q84UV6 SKP1 [Nicotiana benthamiana] 226 1e-58
UniRef100_Q6J4P0 Skp1/Ask1-like protein [Zantedeschia hybrid cul... 226 1e-58
UniRef100_Q852T7 Kinetochore protein [Brassica juncea] 225 4e-58
UniRef100_Q852T4 Kinetochore protein [Brassica juncea] 225 4e-58
UniRef100_O48570 Fimbriata-associated protein [Antirrhinum majus] 224 5e-58
UniRef100_O65283 UIP2 [Arabidopsis thaliana] 224 6e-58
UniRef100_Q852T9 Kinetochore protein [Brassica juncea] 222 3e-57
UniRef100_Q39255 Arabidopsis thaliana Skp1p homolog [Arabidopsis... 221 4e-57
UniRef100_Q852T6 Kinetochore protein [Brassica juncea] 220 9e-57
UniRef100_Q9M3X1 Putative kinetochore protein [Hordeum vulgare] 219 2e-56
UniRef100_Q70JS0 Putative SKP1 protein [Triticum aestivum] 218 3e-56
UniRef100_Q6PL11 Skp1 protein [Oryza sativa] 217 8e-56
UniRef100_Q7XYF8 SKP1/ASK1-like protein [Triticum aestivum] 215 3e-55
UniRef100_Q9LNT9 T20H2.8 protein [Arabidopsis thaliana] 209 3e-53
UniRef100_Q651E8 Putative UIP2 [Oryza sativa] 208 5e-53
>UniRef100_Q9XF99 Skp1 [Medicago sativa]
Length = 153
Score = 253 bits (646), Expect = 1e-66
Identities = 122/153 (79%), Positives = 142/153 (92%), Gaps = 1/153 (0%)
Query: 11 TKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKK 70
T+KITLKSSD ETFE+++AVALESQTIKH+I+D+CAD SGIPLPNVT KILA VIE+CKK
Sbjct: 2 TRKITLKSSDGETFEVDEAVALESQTIKHMIEDDCAD-SGIPLPNVTSKILAKVIEYCKK 60
Query: 71 HVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKG 130
HVDA ++++KP+EDE+ WD+EFVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMIKG
Sbjct: 61 HVDAAAAEDKPNEDELKSWDSEFVKVDQATLFDLILAANYLNIKSLLDLTCQTVADMIKG 120
Query: 131 RTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
+TPEEIRKTFNI ND++PEEEEEVR E QWAF+
Sbjct: 121 KTPEEIRKTFNIKNDFSPEEEEEVRRENQWAFE 153
>UniRef100_Q68GS0 SKP1 [Nicotiana tabacum]
Length = 155
Score = 239 bits (611), Expect = 1e-62
Identities = 118/155 (76%), Positives = 139/155 (89%), Gaps = 1/155 (0%)
Query: 9 TATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHC 68
+++K I LKSSD ETFE+E+AVALESQTIKH+I+D+CAD S IPLPNVT KILA VIE+C
Sbjct: 2 SSSKMIVLKSSDGETFEVEEAVALESQTIKHMIEDDCADTS-IPLPNVTSKILAKVIEYC 60
Query: 69 KKHVDATSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMI 128
K+HVDAT +++K SEDE+ +D++FVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMI
Sbjct: 61 KRHVDATKTEDKASEDELKGFDSDFVKVDQATLFDLILAANYLNIKSLLDLTCQTVADMI 120
Query: 129 KGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
KG+TPEEIRKTFNI ND+TPEEEEEVR E WAF+
Sbjct: 121 KGKTPEEIRKTFNIKNDFTPEEEEEVRRENAWAFE 155
>UniRef100_O82463 SKP1-like protein [Nicotiana clevelandii]
Length = 153
Score = 228 bits (582), Expect = 3e-59
Identities = 114/153 (74%), Positives = 134/153 (87%), Gaps = 2/153 (1%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K I L+SSD ETFE+E++VALESQTIKH+I+D+CAD S IPLPNVT KILA VIE+CK+H
Sbjct: 2 KMIVLRSSDGETFEVEESVALESQTIKHMIEDDCADTS-IPLPNVTSKILAKVIEYCKRH 60
Query: 72 VDATS-SDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKG 130
VDA S +++K ED++ +D +FVKVDQ TLFDLILAANYLNIKSLLDLTC+TVADMIKG
Sbjct: 61 VDAASKTEDKAVEDDLKAFDADFVKVDQSTLFDLILAANYLNIKSLLDLTCQTVADMIKG 120
Query: 131 RTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
+TPEEIRKTFNI ND+TPEEEEEVR E WAF+
Sbjct: 121 KTPEEIRKTFNIKNDFTPEEEEEVRRENAWAFE 153
>UniRef100_O48569 Fimbriata-associated protein [Antirrhinum majus]
Length = 161
Score = 228 bits (580), Expect = 6e-59
Identities = 115/160 (71%), Positives = 139/160 (86%), Gaps = 8/160 (5%)
Query: 11 TKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKK 70
TKKITL+SSD E FE+E+++ALESQTIKH+I+D+CAD+ IPLPNVTGKIL+ VIE+CK+
Sbjct: 3 TKKITLRSSDGEVFEVEESLALESQTIKHMIEDDCADNV-IPLPNVTGKILSKVIEYCKR 61
Query: 71 HVDATSS--DEK-----PSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
HVDA ++ D+K S+DE+ +D +FVKVDQ TLFDLILAANYLNIK+LLDLTC+T
Sbjct: 62 HVDAAAAKADDKLASTGTSDDELKAFDADFVKVDQATLFDLILAANYLNIKTLLDLTCQT 121
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 122 VADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 161
>UniRef100_Q852T5 Kinetochore protein [Brassica juncea]
Length = 160
Score = 226 bits (577), Expect = 1e-58
Identities = 111/160 (69%), Positives = 135/160 (84%), Gaps = 7/160 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD E+FE+++AVALESQTI H+++D+C D+ GIPLPNVT KILA VIE+CK
Sbjct: 2 STKKIVLKSSDGESFEVDEAVALESQTIAHMVEDDCVDN-GIPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEKP------SEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
KHVDA +S + S+D++ WD EF+K+DQ TLF+LILAANYLNIK+LLDLTC+T
Sbjct: 61 KHVDAAASKTEAVDGGASSDDDLKAWDAEFMKIDQATLFELILAANYLNIKNLLDLTCQT 120
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIR TFNI ND+TPEEEEEVR E QWAF+
Sbjct: 121 VADMIKGKTPEEIRTTFNIKNDFTPEEEEEVRRENQWAFE 160
>UniRef100_Q84UV6 SKP1 [Nicotiana benthamiana]
Length = 153
Score = 226 bits (577), Expect = 1e-58
Identities = 113/153 (73%), Positives = 133/153 (86%), Gaps = 2/153 (1%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K I L+SSD ETFE+E++VALESQTIKH+I+D+CAD S IPLPNVT KILA VIE+CK+H
Sbjct: 2 KMIVLRSSDGETFEVEESVALESQTIKHMIEDDCADTS-IPLPNVTSKILAKVIEYCKRH 60
Query: 72 VDATS-SDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTVADMIKG 130
VDA S +++K ED++ +D +FVKVDQ TLFDLILAANYLNIK LLDLTC+TVADMIKG
Sbjct: 61 VDAASKTEDKAVEDDLKAFDADFVKVDQSTLFDLILAANYLNIKRLLDLTCQTVADMIKG 120
Query: 131 RTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
+TPEEIRKTFNI ND+TPEEEEEVR E WAF+
Sbjct: 121 KTPEEIRKTFNIKNDFTPEEEEEVRRENAWAFE 153
>UniRef100_Q6J4P0 Skp1/Ask1-like protein [Zantedeschia hybrid cultivar]
Length = 167
Score = 226 bits (577), Expect = 1e-58
Identities = 114/168 (67%), Positives = 140/168 (82%), Gaps = 6/168 (3%)
Query: 1 MSDGVESSTATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKI 60
M+ ++ A+KK+TLKSSD E FE+E+ VA+ESQTI++LI+D+C D G+PLPNVTG+I
Sbjct: 1 MAADAAAAGASKKLTLKSSDGEVFEVEETVAMESQTIRNLIEDDCTAD-GVPLPNVTGRI 59
Query: 61 LAMVIEHCKKHVD--ATSSDE---KPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKS 115
LA VIE CKKHV+ A +DE + +++E+ WD +FVKVDQ TLFDLILAANYLNIKS
Sbjct: 60 LAKVIEFCKKHVEIAALKADEGVDRAADEELKVWDADFVKVDQTTLFDLILAANYLNIKS 119
Query: 116 LLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
LLDLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 120 LLDLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 167
>UniRef100_Q852T7 Kinetochore protein [Brassica juncea]
Length = 160
Score = 225 bits (573), Expect = 4e-58
Identities = 109/160 (68%), Positives = 135/160 (84%), Gaps = 7/160 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD E+FE+++AVALESQTI H+++D+C D+ G+PLPNVT KILA VIE+CK
Sbjct: 2 STKKIVLKSSDGESFEVDEAVALESQTIAHMVEDDCVDN-GVPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEKP------SEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
KHVDA +S + S+D++ WD EF+K+DQ TLF+LILAANYLNIK+LLDLTC+T
Sbjct: 61 KHVDAVASKSEAVDGGGSSDDDLKAWDAEFMKIDQATLFELILAANYLNIKNLLDLTCQT 120
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIR TFNI ND++PEEEEEVR E QWAF+
Sbjct: 121 VADMIKGKTPEEIRTTFNIKNDFSPEEEEEVRRENQWAFE 160
>UniRef100_Q852T4 Kinetochore protein [Brassica juncea]
Length = 160
Score = 225 bits (573), Expect = 4e-58
Identities = 109/160 (68%), Positives = 135/160 (84%), Gaps = 7/160 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD E+FE+++AVALESQTI H+++D+C D+ G+PLPNVT KILA VIE+CK
Sbjct: 2 STKKIVLKSSDGESFEVDEAVALESQTIAHMVEDDCVDN-GVPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEKP------SEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
KHVDA +S + S+D++ WD EF+K+DQ TLF+LILAANYLNIK+LLDLTC+T
Sbjct: 61 KHVDAAASKSEAVDGGGSSDDDLKAWDAEFMKIDQATLFELILAANYLNIKNLLDLTCQT 120
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIR TFNI ND++PEEEEEVR E QWAF+
Sbjct: 121 VADMIKGKTPEEIRTTFNIKNDFSPEEEEEVRRENQWAFE 160
>UniRef100_O48570 Fimbriata-associated protein [Antirrhinum majus]
Length = 165
Score = 224 bits (572), Expect = 5e-58
Identities = 114/164 (69%), Positives = 140/164 (84%), Gaps = 8/164 (4%)
Query: 7 SSTATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIE 66
S A+KKITL+SSD E FE+++A+AL SQTIKH+I+D+CAD+ IPLPNVTGKIL+ VIE
Sbjct: 3 SDNASKKITLRSSDGEVFEVDEAIALLSQTIKHMIEDDCADNV-IPLPNVTGKILSKVIE 61
Query: 67 HCKKHVDATS--SDEKPS-----EDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDL 119
+CK+HVDA + S+EK + +D++ +D +FVKVDQ TLFDLILAANYLNIK+LLDL
Sbjct: 62 YCKRHVDADAAKSEEKVAAAAAGDDDLKAFDADFVKVDQATLFDLILAANYLNIKTLLDL 121
Query: 120 TCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
TC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 122 TCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 165
>UniRef100_O65283 UIP2 [Arabidopsis thaliana]
Length = 172
Score = 224 bits (571), Expect = 6e-58
Identities = 112/171 (65%), Positives = 134/171 (77%), Gaps = 17/171 (9%)
Query: 9 TATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHC 68
+ +KITLKSSD E FEI++AVALESQTIKH+I+D+C D+ GIPLPNVT KIL+ VIE+C
Sbjct: 3 STVRKITLKSSDGENFEIDEAVALESQTIKHMIEDDCTDN-GIPLPNVTSKILSKVIEYC 61
Query: 69 KKHVDATSSDEKP----------------SEDEINKWDTEFVKVDQDTLFDLILAANYLN 112
K+HV+A E S++++ WD+EF+KVDQ TLFDLILAANYLN
Sbjct: 62 KRHVEAAEKSETTADAAAATTTTTVASGSSDEDLKTWDSEFIKVDQGTLFDLILAANYLN 121
Query: 113 IKSLLDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
IK LLDLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 122 IKGLLDLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 172
>UniRef100_Q852T9 Kinetochore protein [Brassica juncea]
Length = 161
Score = 222 bits (565), Expect = 3e-57
Identities = 107/161 (66%), Positives = 135/161 (83%), Gaps = 8/161 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD+E+FE+++AVA ESQT+ H+++D+C D+ GIPLPNVTGKILA VIE+CK
Sbjct: 2 STKKIVLKSSDDESFEVDEAVARESQTLAHMVEDDCTDN-GIPLPNVTGKILAKVIEYCK 60
Query: 70 KHVDA-------TSSDEKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCK 122
KHVDA T+ PS++++ WD EF+ +DQ TLF+LILAANYLNIK+LLDLTC+
Sbjct: 61 KHVDAAAAKTEATADGGAPSDEDLKAWDAEFMNIDQATLFELILAANYLNIKNLLDLTCQ 120
Query: 123 TVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
TVADMIKG+TP+EIR TFNI ND++PEEEEEVR E QWAF+
Sbjct: 121 TVADMIKGKTPDEIRTTFNIKNDFSPEEEEEVRRENQWAFE 161
>UniRef100_Q39255 Arabidopsis thaliana Skp1p homolog [Arabidopsis thaliana]
Length = 160
Score = 221 bits (564), Expect = 4e-57
Identities = 107/160 (66%), Positives = 134/160 (82%), Gaps = 7/160 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+ KKI LKSSD E+FE+E+AVALESQTI H+++D+C D+ G+PLPNVT KILA VIE+CK
Sbjct: 2 SAKKIVLKSSDGESFEVEEAVALESQTIAHMVEDDCVDN-GVPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEKP------SEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKT 123
+HV+A +S + S+D++ WD +F+K+DQ TLF+LILAANYLNIK+LLDLTC+T
Sbjct: 61 RHVEAAASKAEAVEGAATSDDDLKAWDADFMKIDQATLFELILAANYLNIKNLLDLTCQT 120
Query: 124 VADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
VADMIKG+TPEEIR TFNI ND+TPEEEEEVR E QWAF+
Sbjct: 121 VADMIKGKTPEEIRTTFNIKNDFTPEEEEEVRRENQWAFE 160
>UniRef100_Q852T6 Kinetochore protein [Brassica juncea]
Length = 161
Score = 220 bits (561), Expect = 9e-57
Identities = 107/161 (66%), Positives = 135/161 (83%), Gaps = 8/161 (4%)
Query: 10 ATKKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCK 69
+TKKI LKSSD E+FE+++AVA ESQT+ H+++D+C ++ GIPLPNVT KILA VIE+CK
Sbjct: 2 STKKIVLKSSDGESFEVDEAVARESQTLAHMVEDDCIEN-GIPLPNVTSKILAKVIEYCK 60
Query: 70 KHVDATSSDEK-------PSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCK 122
KHVDA ++ + S+D++ WDTEF+K+DQ TLF+LILAANYLNIK+LLDLTC+
Sbjct: 61 KHVDAAAAKTEGAVDGAASSDDDLKAWDTEFMKIDQATLFELILAANYLNIKNLLDLTCQ 120
Query: 123 TVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
TVADMIKG+TPEEIR TFNI ND++PEEEEEVR E QWAF+
Sbjct: 121 TVADMIKGKTPEEIRTTFNIKNDFSPEEEEEVRRENQWAFE 161
>UniRef100_Q9M3X1 Putative kinetochore protein [Hordeum vulgare]
Length = 175
Score = 219 bits (559), Expect = 2e-56
Identities = 111/166 (66%), Positives = 129/166 (76%), Gaps = 15/166 (9%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K ITLKSSD E FE+E+AVA+ESQTI+H+I+D+CAD+ GIPLPNV KIL+ VIE+C KH
Sbjct: 11 KMITLKSSDGEEFEVEEAVAMESQTIRHMIEDDCADN-GIPLPNVNSKILSKVIEYCNKH 69
Query: 72 VDATSSDEKPSED--------------EINKWDTEFVKVDQDTLFDLILAANYLNIKSLL 117
V A +D S D ++ WD EFVKVDQ TLFDLILAANYLNIK LL
Sbjct: 70 VQAKPADAGASSDTASAAXGAPAAPAEDLKNWDAEFVKVDQATLFDLILAANYLNIKGLL 129
Query: 118 DLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
DLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEE+R E QWAF+
Sbjct: 130 DLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEIRRENQWAFE 175
>UniRef100_Q70JS0 Putative SKP1 protein [Triticum aestivum]
Length = 174
Score = 218 bits (556), Expect = 3e-56
Identities = 112/166 (67%), Positives = 130/166 (77%), Gaps = 16/166 (9%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K ITLKSSD E FE+E+AVA+ESQTI+H+I+D+CAD+ GIPLPNV KIL+ VIE+C KH
Sbjct: 11 KMITLKSSDGEEFEVEEAVAMESQTIRHMIEDDCADN-GIPLPNVNSKILSKVIEYCNKH 69
Query: 72 VDATSSD--------------EKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLL 117
V A +D P+ED + WD EFVKVDQ TLFDLILAANYLNIK LL
Sbjct: 70 VQAKPADGAAAAAGASDAAAPTAPAED-LKNWDAEFVKVDQATLFDLILAANYLNIKGLL 128
Query: 118 DLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
DLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEE+R E QWAF+
Sbjct: 129 DLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEIRRENQWAFE 174
>UniRef100_Q6PL11 Skp1 protein [Oryza sativa]
Length = 173
Score = 217 bits (553), Expect = 8e-56
Identities = 109/167 (65%), Positives = 130/167 (77%), Gaps = 16/167 (9%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K ITLKSSD E FE+E+AVA+ESQTI+H+I+D+CAD+ GIPLPNV KIL+ VIE+C KH
Sbjct: 8 KMITLKSSDGEEFEVEEAVAMESQTIRHMIEDDCADN-GIPLPNVNSKILSKVIEYCNKH 66
Query: 72 VDATSSDEK---------------PSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSL 116
V A ++ PS +++ WD +FVKVDQ TLFDLILAANYLNIK L
Sbjct: 67 VHAAAAAASKAADDAASAAAAVPPPSGEDLKNWDADFVKVDQATLFDLILAANYLNIKGL 126
Query: 117 LDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
LDLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEE+R E QWAF+
Sbjct: 127 LDLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEIRRENQWAFE 173
>UniRef100_Q7XYF8 SKP1/ASK1-like protein [Triticum aestivum]
Length = 175
Score = 215 bits (548), Expect = 3e-55
Identities = 111/167 (66%), Positives = 129/167 (76%), Gaps = 17/167 (10%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K ITLKSSD E FE+E+AVA+ESQTI+H+I+D+CAD+ GIPLPNV KIL+ VIE+C KH
Sbjct: 11 KMITLKSSDGEEFEVEEAVAMESQTIRHMIEDDCADN-GIPLPNVNSKILSKVIEYCNKH 69
Query: 72 VDATSSD---------------EKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSL 116
V A +D P+ED + WD EFVKVDQ TLFDLILAANYLNIK L
Sbjct: 70 VQAKPADGAAAGAGAGASDAAPAAPAED-LKNWDAEFVKVDQATLFDLILAANYLNIKGL 128
Query: 117 LDLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
DLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEE+R E QWAF+
Sbjct: 129 PDLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEIRRENQWAFE 175
>UniRef100_Q9LNT9 T20H2.8 protein [Arabidopsis thaliana]
Length = 163
Score = 209 bits (531), Expect = 3e-53
Identities = 105/159 (66%), Positives = 127/159 (79%), Gaps = 8/159 (5%)
Query: 12 KKITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKH 71
K I LKSSD E+FEIE+AVA++SQTIKH+I+D+CAD+ GIPLPNVTG ILA VIE+CKKH
Sbjct: 6 KMIILKSSDGESFEIEEAVAVKSQTIKHMIEDDCADN-GIPLPNVTGAILAKVIEYCKKH 64
Query: 72 VDATSSDEKPSE-------DEINKWDTEFVKVDQDTLFDLILAANYLNIKSLLDLTCKTV 124
V+A + + DE+ WD+EFVKVDQ TLFDLILAANYLNI LLDLTCK V
Sbjct: 65 VEAAAEAGGDKDFYGSAENDELKNWDSEFVKVDQPTLFDLILAANYLNIGGLLDLTCKAV 124
Query: 125 ADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
AD ++G+TPE++R FNI NDYTPEEE EVR+E +WAF+
Sbjct: 125 ADQMRGKTPEQMRAHFNIKNDYTPEEEAEVRNENKWAFE 163
>UniRef100_Q651E8 Putative UIP2 [Oryza sativa]
Length = 175
Score = 208 bits (529), Expect = 5e-53
Identities = 106/166 (63%), Positives = 126/166 (75%), Gaps = 17/166 (10%)
Query: 14 ITLKSSDNETFEIEKAVALESQTIKHLIDDNCADDSGIPLPNVTGKILAMVIEHCKKHVD 73
ITL+S + + FE+ +AVA+ESQTI+H+I+D CAD +GIPLPNV+ KIL+ VIE+C KHV+
Sbjct: 11 ITLRSCEGQVFEVAEAVAMESQTIRHMIEDKCAD-TGIPLPNVSAKILSKVIEYCSKHVE 69
Query: 74 ATSSD----------------EKPSEDEINKWDTEFVKVDQDTLFDLILAANYLNIKSLL 117
A K EDE+ +D EFVKVDQ TLFDLILAANYLNIK LL
Sbjct: 70 ARGGAAAAADGDAPAPAAVEANKAVEDELKTFDAEFVKVDQSTLFDLILAANYLNIKGLL 129
Query: 118 DLTCKTVADMIKGRTPEEIRKTFNIINDYTPEEEEEVRSETQWAFD 163
DLTC+TVADMIKG+TPEEIRKTFNI ND+TPEEEEEVR E QWAF+
Sbjct: 130 DLTCQTVADMIKGKTPEEIRKTFNIKNDFTPEEEEEVRRENQWAFE 175
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.310 0.129 0.358
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 267,923,650
Number of Sequences: 2790947
Number of extensions: 10487859
Number of successful extensions: 27148
Number of sequences better than 10.0: 238
Number of HSP's better than 10.0 without gapping: 191
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 26679
Number of HSP's gapped (non-prelim): 245
length of query: 164
length of database: 848,049,833
effective HSP length: 117
effective length of query: 47
effective length of database: 521,509,034
effective search space: 24510924598
effective search space used: 24510924598
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC148341.11


