

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC147472.6 + phase: 0 /pseudo
(180 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
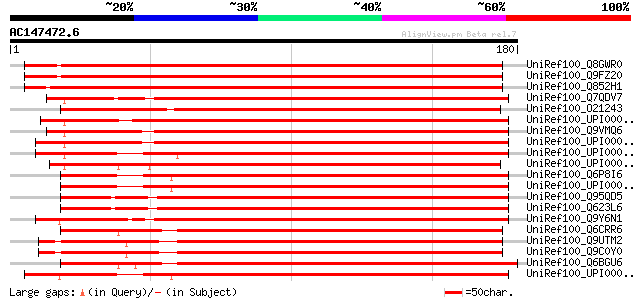
UniRef100_Q8GWR0 Hypothetical protein At1g02410/T6A9_19 [Arabido... 282 2e-75
UniRef100_Q9FZ20 T6A9.10 protein [Arabidopsis thaliana] 282 2e-75
UniRef100_Q852H1 Putative cytochrome c oxidase assembly protein ... 270 2e-71
UniRef100_Q7QDV7 ENSANGP00000022656 [Anopheles gambiae str. PEST] 186 2e-46
UniRef100_O21243 Cytochrome c oxidase assembly protein ctaG [Rec... 186 2e-46
UniRef100_UPI00004329F3 UPI00004329F3 UniRef100 entry 182 3e-45
UniRef100_Q9VMQ6 CG31648-PA [Drosophila melanogaster] 178 7e-44
UniRef100_UPI000034DD0C UPI000034DD0C UniRef100 entry 177 1e-43
UniRef100_UPI00000187D7 UPI00000187D7 UniRef100 entry 176 2e-43
UniRef100_UPI00003C128D UPI00003C128D UniRef100 entry 174 1e-42
UniRef100_Q6P8I6 COX11 homolog, cytochrome c oxidase assembly pr... 170 1e-41
UniRef100_UPI000017FB81 UPI000017FB81 UniRef100 entry 169 3e-41
UniRef100_Q95QD5 Hypothetical protein JC8.5 [Caenorhabditis eleg... 169 3e-41
UniRef100_Q623L6 Hypothetical protein CBG01804 [Caenorhabditis b... 169 4e-41
UniRef100_Q9Y6N1 Cytochrome c oxidase assembly protein COX11, mi... 168 5e-41
UniRef100_Q6CRR6 Kluyveromyces lactis strain NRRL Y-1140 chromos... 167 9e-41
UniRef100_Q9UTM2 Cox1101 protein [Schizosaccharomyces pombe] 166 2e-40
UniRef100_Q9C0Y0 Cox11-b protein [Schizosaccharomyces pombe] 166 2e-40
UniRef100_Q6BGU6 Similar to CA3958|CaCOX11 Candida albicans CaCO... 166 2e-40
UniRef100_UPI000036D5BF UPI000036D5BF UniRef100 entry 164 1e-39
>UniRef100_Q8GWR0 Hypothetical protein At1g02410/T6A9_19 [Arabidopsis thaliana]
Length = 287
Score = 282 bits (722), Expect = 2e-75
Identities = 137/170 (80%), Positives = 149/170 (87%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
LT + FGM LT +A+ PLYRTFCQAT Y GTV R+E VEEKIARH + TVT REIVVQ
Sbjct: 97 LTAVVFGMVGLT-YAAVPLYRTFCQATGYGGTVQRKETVEEKIARHSESGTVTEREIVVQ 155
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGM WKF PTQREVRVKPGESALAFYT EN+SS PITGVSTYNVTPMK GVYFN
Sbjct: 156 FNADVADGMQWKFTPTQREVRVKPGESALAFYTAENKSSAPITGVSTYNVTPMKAGVYFN 215
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQRLLPGE+IDMPVFFYIDPE E DP+M+GINN+ILSYTFFK
Sbjct: 216 KIQCFCFEEQRLLPGEQIDMPVFFYIDPEFETDPRMDGINNLILSYTFFK 265
>UniRef100_Q9FZ20 T6A9.10 protein [Arabidopsis thaliana]
Length = 216
Score = 282 bits (722), Expect = 2e-75
Identities = 137/170 (80%), Positives = 149/170 (87%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
LT + FGM LT +A+ PLYRTFCQAT Y GTV R+E VEEKIARH + TVT REIVVQ
Sbjct: 26 LTAVVFGMVGLT-YAAVPLYRTFCQATGYGGTVQRKETVEEKIARHSESGTVTEREIVVQ 84
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGM WKF PTQREVRVKPGESALAFYT EN+SS PITGVSTYNVTPMK GVYFN
Sbjct: 85 FNADVADGMQWKFTPTQREVRVKPGESALAFYTAENKSSAPITGVSTYNVTPMKAGVYFN 144
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQRLLPGE+IDMPVFFYIDPE E DP+M+GINN+ILSYTFFK
Sbjct: 145 KIQCFCFEEQRLLPGEQIDMPVFFYIDPEFETDPRMDGINNLILSYTFFK 194
>UniRef100_Q852H1 Putative cytochrome c oxidase assembly protein [Oryza sativa]
Length = 244
Score = 270 bits (689), Expect = 2e-71
Identities = 129/170 (75%), Positives = 144/170 (83%), Gaps = 1/170 (0%)
Query: 6 LTGLAFGMGVLTIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQ 65
L G+A M V +A+ PLYR FCQAT Y GTV RRE VEEKI+RH + T TSREI+VQ
Sbjct: 72 LLGVAAAM-VGASYAAVPLYRRFCQATGYGGTVQRRESVEEKISRHARDGTTTSREIIVQ 130
Query: 66 FNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFN 125
FNAD++DGMPWKF+PTQREV+VKPGESALAFYT EN+SS PITGVSTYNV PMK +YFN
Sbjct: 131 FNADVADGMPWKFIPTQREVKVKPGESALAFYTAENRSSAPITGVSTYNVAPMKAAIYFN 190
Query: 126 KIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
KIQCFCFEEQ LLPGE+IDMPVFFYIDPE E DPKM G+NNI+LSYTFFK
Sbjct: 191 KIQCFCFEEQTLLPGEQIDMPVFFYIDPEFETDPKMEGVNNIVLSYTFFK 240
>UniRef100_Q7QDV7 ENSANGP00000022656 [Anopheles gambiae str. PEST]
Length = 195
Score = 186 bits (473), Expect = 2e-46
Identities = 93/167 (55%), Positives = 119/167 (70%), Gaps = 7/167 (4%)
Query: 14 GVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADI 70
GVLT+ +A+ PLYR FCQA SY GT T + EK+ +S Q V R I ++FNADI
Sbjct: 19 GVLTVGLSYAAVPLYRMFCQAYSYGGT-TSQGHDGEKV---ESMQKVKDRVIKIKFNADI 74
Query: 71 SDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCF 130
M W F P Q E+RV PGE+ALAFY+ +N + P+ G+STYNV P + G YFNKIQCF
Sbjct: 75 GASMRWNFKPQQTEIRVVPGETALAFYSAKNPTDHPVVGISTYNVIPFEAGAYFNKIQCF 134
Query: 131 CFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFEEQ+L P E++D+PVFFYIDP+ +DPKM +++I LSYTFF+ K
Sbjct: 135 CFEEQQLNPHEEVDLPVFFYIDPDFIEDPKMELVDSITLSYTFFESK 181
>UniRef100_O21243 Cytochrome c oxidase assembly protein ctaG [Reclinomonas americana]
Length = 182
Score = 186 bits (473), Expect = 2e-46
Identities = 89/156 (57%), Positives = 111/156 (71%), Gaps = 2/156 (1%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
+ S PLYR FCQ T + GT + + + D Q +R I V+FN D+SD MPWKF
Sbjct: 24 YGSVPLYRIFCQVTGFGGTTQVADLESDILTLKDEQQE--NRIITVRFNGDVSDTMPWKF 81
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q+E++V GE+ALAFY+ EN + + I G+STYNV P + G+YFNKIQCFCFEEQRL
Sbjct: 82 HPIQQEIKVMVGETALAFYSAENPTDSSIIGISTYNVNPQQAGIYFNKIQCFCFEEQRLK 141
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFF 174
P E IDMPVFF+IDP I DDPKM+ I++I LSYTFF
Sbjct: 142 PHETIDMPVFFFIDPAILDDPKMSDIDSITLSYTFF 177
>UniRef100_UPI00004329F3 UPI00004329F3 UniRef100 entry
Length = 195
Score = 182 bits (462), Expect = 3e-45
Identities = 90/169 (53%), Positives = 113/169 (66%), Gaps = 7/169 (4%)
Query: 12 GMGVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
G G+L I +AS PLYR FCQ+ +Y GT++ V + S + + R I ++FNA
Sbjct: 18 GFGILVIGFTYASVPLYRIFCQSYNYGGTLS----VNHDNTKVQSMKPIKDRVIKIKFNA 73
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
DI M W F P Q ++V PGE+ALAFY +N PITG+STYNV P + YFNKIQ
Sbjct: 74 DIGAMMQWNFKPQQNFIKVMPGETALAFYRAQNPLDIPITGISTYNVVPYEAAQYFNKIQ 133
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DPKM + I+LSYTFF+ K
Sbjct: 134 CFCFEEQRLNPHEEVDMPVFFYIDPEFVNDPKMEDVEEIVLSYTFFEAK 182
>UniRef100_Q9VMQ6 CG31648-PA [Drosophila melanogaster]
Length = 241
Score = 178 bits (451), Expect = 7e-44
Identities = 87/167 (52%), Positives = 111/167 (66%), Gaps = 7/167 (4%)
Query: 14 GVLTI---FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADI 70
GVL + +A+ PLY FCQA SY GT T+ E+ + + + R + ++FNADI
Sbjct: 61 GVLIVGLSYAAVPLYSIFCQAYSYGGTTTQGHDAEKV----EHMKKIEDRVLKIRFNADI 116
Query: 71 SDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCF 130
M W F P Q E++V PGE+ALAFYT N + P+ G+STYNV P + G YFNKIQCF
Sbjct: 117 GSSMRWNFKPQQYEIKVAPGETALAFYTARNPTDKPVIGISTYNVIPFEAGAYFNKIQCF 176
Query: 131 CFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFEEQ+L P E++DMPVFFYIDPEI DP + + I LSYTFF+ K
Sbjct: 177 CFEEQQLNPHEEVDMPVFFYIDPEITADPALETCDTITLSYTFFEAK 223
>UniRef100_UPI000034DD0C UPI000034DD0C UniRef100 entry
Length = 263
Score = 177 bits (449), Expect = 1e-43
Identities = 88/169 (52%), Positives = 114/169 (67%), Gaps = 5/169 (2%)
Query: 10 AFGMGVLTI-FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A G+G++ + +A+ PLYR +CQA+ GT ++ ++ + V R I V FNA
Sbjct: 88 AAGVGMIGLSYAAVPLYRLYCQASGLGGTAVAGHNADQV----ETMKPVPERVIKVTFNA 143
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D M W F P Q EV V PGE+ALAFY +N + PITG+STYNV P + G YFNKIQ
Sbjct: 144 DRHASMQWNFRPQQTEVFVVPGETALAFYKAKNPTDKPITGISTYNVVPFEAGQYFNKIQ 203
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE ++DP+M ++ I LSYTFF+ K
Sbjct: 204 CFCFEEQRLNPHEEVDMPVFFYIDPEFDEDPRMARVDTITLSYTFFEAK 252
>UniRef100_UPI00000187D7 UPI00000187D7 UniRef100 entry
Length = 205
Score = 176 bits (447), Expect = 2e-43
Identities = 90/174 (51%), Positives = 115/174 (65%), Gaps = 15/174 (8%)
Query: 10 AFGMGVLTI-FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVT-----SREIV 63
A G+G++ + +A+ PLYR +CQA+ G +A HD++Q T R I
Sbjct: 30 AAGVGMIGLSYAAVPLYRLYCQASGLGGMA---------VAGHDADQVETMKPVPERVIK 80
Query: 64 VQFNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVY 123
+ FNAD M W F P Q EV V PGE+ALAFY +N + PITG+STYNV P + G Y
Sbjct: 81 ITFNADRHASMQWNFRPQQTEVFVVPGETALAFYRAKNPTDKPITGISTYNVVPFEAGQY 140
Query: 124 FNKIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
FNKIQCFCFEEQRL P E++DMPVFFYIDPE ++DP+M ++ I LSYTFF+ K
Sbjct: 141 FNKIQCFCFEEQRLNPHEEVDMPVFFYIDPEFDEDPRMARVDTITLSYTFFEAK 194
>UniRef100_UPI00003C128D UPI00003C128D UniRef100 entry
Length = 349
Score = 174 bits (440), Expect = 1e-42
Identities = 83/167 (49%), Positives = 116/167 (68%), Gaps = 7/167 (4%)
Query: 15 VLTI---FASPPLYRTFCQATSYDGT-VTRRERVEEKI---ARHDSNQTVTSREIVVQFN 67
VLT+ +A+ PLYR FC AT + GT +T ER A D + ++ I V FN
Sbjct: 157 VLTLGFSYAAVPLYRAFCSATGFSGTPITDPERFAPSRLVPAYLDPSTNAPTKRIRVTFN 216
Query: 68 ADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKI 127
AD SD +PW F P Q+E+ V PGE+ALAFYT +N+S I G++TYNV+P ++ YF K+
Sbjct: 217 ADASDAIPWSFTPVQKEIYVLPGETALAFYTAKNKSDKDIIGIATYNVSPDRIAPYFAKV 276
Query: 128 QCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFF 174
+CFCFEEQ+LL GE++D+PVFF+ID ++ DDP G+++++LSYTFF
Sbjct: 277 ECFCFEEQKLLAGEEVDLPVFFFIDSDVLDDPSTKGVDDVVLSYTFF 323
>UniRef100_Q6P8I6 COX11 homolog, cytochrome c oxidase assembly protein [Mus musculus]
Length = 275
Score = 170 bits (431), Expect = 1e-41
Identities = 84/164 (51%), Positives = 106/164 (64%), Gaps = 14/164 (8%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQT-----VTSREIVVQFNADISDG 73
+A+ PLYR +CQ T G+ +A H S+Q V R I V FNAD+
Sbjct: 110 YAAVPLYRLYCQTTGLGGSA---------VAGHSSDQIENMVPVKDRVIKVTFNADVHAS 160
Query: 74 MPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFE 133
+ W F P Q E+ V PGE+ALAFY +N + P+ G+STYNV P + G YFNKIQCFCFE
Sbjct: 161 LQWNFRPQQTEIYVVPGETALAFYKAKNPTDKPVIGISTYNVVPFEAGQYFNKIQCFCFE 220
Query: 134 EQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
EQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 221 EQRLNPQEEVDMPVFFYIDPEFAEDPRMVNVDLITLSYTFFEAK 264
>UniRef100_UPI000017FB81 UPI000017FB81 UniRef100 entry
Length = 275
Score = 169 bits (428), Expect = 3e-41
Identities = 84/164 (51%), Positives = 106/164 (64%), Gaps = 14/164 (8%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQT-----VTSREIVVQFNADISDG 73
+A+ PLYR +CQ T G+ +A H S+Q V R I V FNAD+
Sbjct: 110 YAAVPLYRLYCQTTGLGGSA---------VAGHASDQIENMVPVKDRIIKVTFNADVHAS 160
Query: 74 MPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFE 133
+ W F P Q E+ V PGE+ALAFY +N + P+ G+STYNV P + G YFNKIQCFCFE
Sbjct: 161 LQWNFRPQQTEIYVVPGETALAFYKAKNPTDKPVIGISTYNVVPFEAGQYFNKIQCFCFE 220
Query: 134 EQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
EQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 221 EQRLNPQEEVDMPVFFYIDPEFAEDPRMVNVDLITLSYTFFEAK 264
>UniRef100_Q95QD5 Hypothetical protein JC8.5 [Caenorhabditis elegans]
Length = 195
Score = 169 bits (428), Expect = 3e-41
Identities = 82/159 (51%), Positives = 112/159 (69%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
FA+ P YR FC+ TS+ G +T+ + +KIA + + R I VQFN+D+ M W+F
Sbjct: 12 FAAIPAYRIFCEQTSFGG-LTQVAKDFDKIA---NMKKCEDRLIRVQFNSDVPSSMRWEF 67
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFYT N + PI G+S+YN+TP + YFNKIQCFCFEEQ L
Sbjct: 68 KPQQHEIYVHPGETALAFYTARNPTDKPIIGISSYNLTPFQAAYYFNKIQCFCFEEQILN 127
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
PGE++D+PVFFYIDP+ +DP + +++I+LSYTFF+ K
Sbjct: 128 PGEQVDLPVFFYIDPDYVNDPALEYLDSILLSYTFFEAK 166
>UniRef100_Q623L6 Hypothetical protein CBG01804 [Caenorhabditis briggsae]
Length = 426
Score = 169 bits (427), Expect = 4e-41
Identities = 82/159 (51%), Positives = 112/159 (69%), Gaps = 4/159 (2%)
Query: 19 FASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPWKF 78
FA+ P YR FC+ TS+ G +T+ + +KIA + + R I VQFN+D+ M W+F
Sbjct: 243 FAAIPAYRIFCEQTSFGG-LTQVAKDFDKIA---NMKKCEDRLIRVQFNSDVPSSMRWEF 298
Query: 79 LPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQRLL 138
P Q E+ V PGE+ALAFYT N + PI G+S+YN+TP + YFNKIQCFCFEEQ L
Sbjct: 299 KPQQHEIFVHPGETALAFYTARNPTDKPIIGISSYNLTPFQAAYYFNKIQCFCFEEQILN 358
Query: 139 PGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
PGE++D+PVFFYIDP+ +DP + +++I+LSYTFF+ K
Sbjct: 359 PGEQVDLPVFFYIDPDYVNDPALEYLDSILLSYTFFEAK 397
>UniRef100_Q9Y6N1 Cytochrome c oxidase assembly protein COX11, mitochondrial
precursor [Homo sapiens]
Length = 276
Score = 168 bits (426), Expect = 5e-41
Identities = 84/169 (49%), Positives = 111/169 (64%), Gaps = 5/169 (2%)
Query: 10 AFGMGVL-TIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQTVTSREIVVQFNA 68
A +G+L +A+ PLYR +CQ T G+ +KI ++ V R I + FNA
Sbjct: 101 AVAVGMLGASYAAVPLYRLYCQTTGLGGSAVAGH-ASDKI---ENMVPVKDRIIKISFNA 156
Query: 69 DISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQ 128
D+ + W F P Q E+ V PGE+ALAFY V+N + P+ G+STYN+ P + G YFNKIQ
Sbjct: 157 DVHASLQWNFRPQQTEIYVVPGETALAFYRVKNPTDKPVIGISTYNIVPFEAGQYFNKIQ 216
Query: 129 CFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
CFCFEEQRL P E++DMPVFFYIDPE +DP+M ++ I LSYTFF+ K
Sbjct: 217 CFCFEEQRLNPQEEVDMPVFFYIDPEFAEDPRMIKVDLITLSYTFFEAK 265
>UniRef100_Q6CRR6 Kluyveromyces lactis strain NRRL Y-1140 chromosome D of strain NRRL
Y- 1140 of Kluyveromyces lactis [Kluyveromyces lactis]
Length = 261
Score = 167 bits (424), Expect = 9e-41
Identities = 80/159 (50%), Positives = 108/159 (67%), Gaps = 7/159 (4%)
Query: 19 FASPPLYRTFCQATSYDGT--VTRRERVEEKIARHDSNQTVTSREIVVQFNADISDGMPW 76
+A+ PLYR C T Y G +R+ ++K+ DSN + I + F +++S +PW
Sbjct: 86 YAAVPLYRAICARTGYGGIPITDKRKFTDDKLIPVDSN-----KRIRISFTSEVSQILPW 140
Query: 77 KFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQR 136
KF+P QREV V PGE+ALAFY +N S I G++TY++TP + YFNKIQCFCFEEQ+
Sbjct: 141 KFVPQQREVYVLPGETALAFYKAKNNSDKDIIGMATYSITPGESSPYFNKIQCFCFEEQK 200
Query: 137 LLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
L GE++DMPVFF+IDP+ DP M I++IIL YTFFK
Sbjct: 201 LAAGEEVDMPVFFFIDPDFASDPSMRNIDDIILHYTFFK 239
>UniRef100_Q9UTM2 Cox1101 protein [Schizosaccharomyces pombe]
Length = 695
Score = 166 bits (421), Expect = 2e-40
Identities = 82/168 (48%), Positives = 108/168 (63%), Gaps = 11/168 (6%)
Query: 11 FGMGVLTIFASPPLYRTFCQATSYDGTVTR---RERVEEKIARHDSNQTVTSREIVVQFN 67
F +G+ +A+ PLYR FC T Y GT+ R E + R D+ + I V FN
Sbjct: 522 FALGLT--YAAVPLYRLFCSKTGYGGTLNTDQSRMNAERMVPRKDN------KRIRVTFN 573
Query: 68 ADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKI 127
D++ + WK P QRE+ V PGE+AL FYT EN S I GV+TYN+ P + VYF+K+
Sbjct: 574 GDVAGNLSWKLWPQQREIYVLPGETALGFYTAENTSDHDIVGVATYNIVPGQAAVYFSKV 633
Query: 128 QCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
CFCFEEQ+L EK+D+PVFF+IDPE DDP M I++I+LSYTFF+
Sbjct: 634 ACFCFEEQKLDAHEKVDLPVFFFIDPEFADDPNMKDIDDILLSYTFFE 681
>UniRef100_Q9C0Y0 Cox11-b protein [Schizosaccharomyces pombe]
Length = 732
Score = 166 bits (421), Expect = 2e-40
Identities = 82/168 (48%), Positives = 108/168 (63%), Gaps = 11/168 (6%)
Query: 11 FGMGVLTIFASPPLYRTFCQATSYDGTVTR---RERVEEKIARHDSNQTVTSREIVVQFN 67
F +G+ +A+ PLYR FC T Y GT+ R E + R D+ + I V FN
Sbjct: 559 FALGLT--YAAVPLYRLFCSKTGYGGTLNTDQSRMNAERMVPRKDN------KRIRVTFN 610
Query: 68 ADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKI 127
D++ + WK P QRE+ V PGE+AL FYT EN S I GV+TYN+ P + VYF+K+
Sbjct: 611 GDVAGNLSWKLWPQQREIYVLPGETALGFYTAENTSDHDIVGVATYNIVPGQAAVYFSKV 670
Query: 128 QCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFK 175
CFCFEEQ+L EK+D+PVFF+IDPE DDP M I++I+LSYTFF+
Sbjct: 671 ACFCFEEQKLDAHEKVDLPVFFFIDPEFADDPNMKDIDDILLSYTFFE 718
>UniRef100_Q6BGU6 Similar to CA3958|CaCOX11 Candida albicans CaCOX11 [Debaryomyces
hansenii]
Length = 261
Score = 166 bits (421), Expect = 2e-40
Identities = 83/164 (50%), Positives = 109/164 (65%), Gaps = 7/164 (4%)
Query: 19 FASPPLYRTFCQATSYDGT-VTRRER-VEEKIARHDSNQTVTSREIVVQFNADISDGMPW 76
FA+ PLYR CQ T Y GT +T R +K+ D+N + I +QF S +PW
Sbjct: 85 FAAVPLYRAICQRTGYGGTPITDSTRFTPDKLIPVDTN-----KRIRIQFTCQASGILPW 139
Query: 77 KFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMKLGVYFNKIQCFCFEEQR 136
KF P QREV V PGE+ALAFY +N S I G++TY+VTP + YFNKIQCFCFEEQR
Sbjct: 140 KFTPQQREVYVVPGETALAFYKAKNMSKEDIIGMATYSVTPEVVAPYFNKIQCFCFEEQR 199
Query: 137 LLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDKVSE 180
L GE++DMPVFF+IDP+ DP+M I +++L+Y+FF+ S+
Sbjct: 200 LSAGEEVDMPVFFFIDPDFAKDPQMRNIEDVVLNYSFFRASYSD 243
>UniRef100_UPI000036D5BF UPI000036D5BF UniRef100 entry
Length = 273
Score = 164 bits (415), Expect = 1e-39
Identities = 84/178 (47%), Positives = 111/178 (62%), Gaps = 15/178 (8%)
Query: 6 LTGLAFGMGVL-TIFASPPLYRTFCQATSYDGTVTRRERVEEKIARHDSNQT-----VTS 59
LT A +G+L +A+ PLYR +CQ T G+ ++ H S+Q V
Sbjct: 94 LTVAAVAVGMLGASYAAVPLYRLYCQTTGLGGSA---------VSGHASDQIENMVPVKD 144
Query: 60 REIVVQFNADISDGMPWKFLPTQREVRVKPGESALAFYTVENQSSTPITGVSTYNVTPMK 119
R I + F+AD+ + W F P Q E+ V PG++AL FY +N + P+ G+STYNV P +
Sbjct: 145 RIIKINFDADVHASLQWNFRPQQTEIYVVPGDTALGFYRAKNPADKPLIGISTYNVVPFE 204
Query: 120 LGVYFNKIQCFCFEEQRLLPGEKIDMPVFFYIDPEIEDDPKMNGINNIILSYTFFKDK 177
G YFNKIQCFCFEEQRL P E++DMPVFFYIDPE DP+M ++ I LSYTFF+ K
Sbjct: 205 AGQYFNKIQCFCFEEQRLNPQEEVDMPVFFYIDPEFAKDPRMINVDLITLSYTFFEAK 262
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.322 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 298,453,816
Number of Sequences: 2790947
Number of extensions: 11533564
Number of successful extensions: 29591
Number of sequences better than 10.0: 383
Number of HSP's better than 10.0 without gapping: 347
Number of HSP's successfully gapped in prelim test: 36
Number of HSP's that attempted gapping in prelim test: 28986
Number of HSP's gapped (non-prelim): 385
length of query: 180
length of database: 848,049,833
effective HSP length: 119
effective length of query: 61
effective length of database: 515,927,140
effective search space: 31471555540
effective search space used: 31471555540
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC147472.6


