

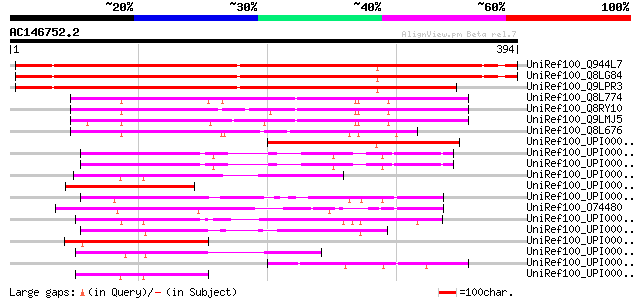
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146752.2 + phase: 0
(394 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q944L7 At1g18480/F15H18_1 [Arabidopsis thaliana] 481 e-134
UniRef100_Q8LG84 Hypothetical protein [Arabidopsis thaliana] 479 e-134
UniRef100_Q9LPR3 F15H18.4 [Arabidopsis thaliana] 459 e-128
UniRef100_Q8L774 Hypothetical protein At1g07010 [Arabidopsis tha... 177 6e-43
UniRef100_Q8RY10 At1g07010/F10K1_19 [Arabidopsis thaliana] 173 7e-42
UniRef100_Q9LMJ5 F10K1.27 protein [Arabidopsis thaliana] 171 3e-41
UniRef100_Q8L676 Hypothetical protein OSJNBa0011L09.21 [Oryza sa... 166 1e-39
UniRef100_UPI00002D058A UPI00002D058A UniRef100 entry 141 4e-32
UniRef100_UPI000042F2AE UPI000042F2AE UniRef100 entry 117 7e-25
UniRef100_UPI0000452C3B UPI0000452C3B UniRef100 entry 115 2e-24
UniRef100_UPI0000280CED UPI0000280CED UniRef100 entry 107 6e-22
UniRef100_UPI000034C07C UPI000034C07C UniRef100 entry 101 4e-20
UniRef100_UPI00003C1C90 UPI00003C1C90 UniRef100 entry 100 7e-20
UniRef100_O74480 SPCC1840.07c protein [Schizosaccharomyces pombe] 100 9e-20
UniRef100_UPI00002F6C5D UPI00002F6C5D UniRef100 entry 99 3e-19
UniRef100_UPI00002B5187 UPI00002B5187 UniRef100 entry 98 5e-19
UniRef100_UPI000042F3C1 UPI000042F3C1 UniRef100 entry 95 4e-18
UniRef100_UPI000030DC8C UPI000030DC8C UniRef100 entry 88 4e-16
UniRef100_UPI00002B8E83 UPI00002B8E83 UniRef100 entry 84 5e-15
UniRef100_UPI00002C3AD7 UPI00002C3AD7 UniRef100 entry 77 8e-13
>UniRef100_Q944L7 At1g18480/F15H18_1 [Arabidopsis thaliana]
Length = 391
Score = 481 bits (1238), Expect = e-134
Identities = 246/395 (62%), Positives = 298/395 (75%), Gaps = 12/395 (3%)
Query: 5 KQNSNTFCNQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSPPPI-PTRLPSPSRLIAIGDL 63
++N + C IP +SSF+DTFVD+SVSG +FLP PSS I TR P RL+AIGDL
Sbjct: 4 RENPSGICKSIPKLISSFVDTFVDYSVSG-IFLPQDPSSQNEILQTRFEKPERLVAIGDL 62
Query: 64 HGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEKLKRQAAIH 123
HGDL+KS+EA IAGLIDSS +TGGS VVQ+GDVLDRGG+E+KILY LEKLKR+A
Sbjct: 63 HGDLEKSREAFKIAGLIDSSDRWTGGSTMVVQVGDVLDRGGEELKILYFLEKLKREAERA 122
Query: 124 GGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLE 183
GG +TMNGNHEIMN EGDFR+ TK G+EEF++W +W+ GNKMK LC GL++ DP E
Sbjct: 123 GGKILTMNGNHEIMNIEGDFRYVTKKGLEEFQIWADWYCLGNKMKTLCSGLDKP-KDPYE 181
Query: 184 NVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVD 243
+ ++F +R + +G RAR+AALRP+GPI+KRF T+N TV VVGDS+FVHGGLL EH++
Sbjct: 182 GIPMSFPRMRADCFEGIRARIAALRPDGPIAKRFLTKNQTVAVVGDSVFVHGGLLAEHIE 241
Query: 244 YGLEKINGEVSDWYKGLFGNRFSPPYCRGRNALVWLRKFSD---GNCDCSSLEHVLSTIP 300
YGLE+IN EV W G G R++P YCRG N++VWLRKFS+ CDC++LEH LSTIP
Sbjct: 242 YGLERINEEVRGWINGFKGGRYAPAYCRGGNSVVWLRKFSEEMAHKCDCAALEHALSTIP 301
Query: 301 GVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEIDR-YGVRILTSNPLY 359
GVKRMIMGHTIQ GINGVC +KAIRIDVGMSKGC GLPEVLEI R GVRI+TSNPLY
Sbjct: 302 GVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPEVLEIRRDSGVRIVTSNPLY 361
Query: 360 NQMNKENVDIGKVEEGFGLLLNNQDGRPRQVEVKA 394
+ +V + G GLL+ P+QVEVKA
Sbjct: 362 KENLYSHV-APDSKTGLGLLV----PVPKQVEVKA 391
>UniRef100_Q8LG84 Hypothetical protein [Arabidopsis thaliana]
Length = 391
Score = 479 bits (1233), Expect = e-134
Identities = 245/395 (62%), Positives = 297/395 (75%), Gaps = 12/395 (3%)
Query: 5 KQNSNTFCNQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSPPPI-PTRLPSPSRLIAIGDL 63
++N + C IP +S F+DTFVD+SVSG +FLP PSS I TR P RL+AIGDL
Sbjct: 4 RENPSGICKSIPKLISYFVDTFVDYSVSG-IFLPQDPSSQNEILQTRFEKPERLVAIGDL 62
Query: 64 HGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEKLKRQAAIH 123
HGDL+KS+EA IAGLIDSS +TGGS VVQ+GDVLDRGG+E+KILY LEKLKR+A
Sbjct: 63 HGDLEKSREAFKIAGLIDSSDRWTGGSTMVVQVGDVLDRGGEELKILYFLEKLKREAERA 122
Query: 124 GGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLE 183
GG +TMNGNHEIMN EGDFR+ TK G+EEF++W +W+ GNKMK LC GL++ DP E
Sbjct: 123 GGKILTMNGNHEIMNIEGDFRYVTKKGLEEFQIWADWYCLGNKMKTLCSGLDKP-KDPYE 181
Query: 184 NVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVD 243
+ ++F +R + +G RAR+AALRP+GPI+KRF T+N TV VVGDS+FVHGGLL EH++
Sbjct: 182 GIPMSFPRMRADCFEGIRARIAALRPDGPIAKRFLTKNQTVAVVGDSVFVHGGLLAEHIE 241
Query: 244 YGLEKINGEVSDWYKGLFGNRFSPPYCRGRNALVWLRKFSD---GNCDCSSLEHVLSTIP 300
YGLE+IN EV W G G R++P YCRG N++VWLRKFS+ CDC++LEH LSTIP
Sbjct: 242 YGLERINEEVRGWINGFKGGRYAPAYCRGGNSVVWLRKFSEEMAHKCDCAALEHALSTIP 301
Query: 301 GVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEIDR-YGVRILTSNPLY 359
GVKRMIMGHTIQ GINGVC +KAIRIDVGMSKGC GLPEVLEI R GVRI+TSNPLY
Sbjct: 302 GVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPEVLEIRRDSGVRIVTSNPLY 361
Query: 360 NQMNKENVDIGKVEEGFGLLLNNQDGRPRQVEVKA 394
+ +V + G GLL+ P+QVEVKA
Sbjct: 362 KENPYSHV-APDSKTGLGLLV----PVPKQVEVKA 391
>UniRef100_Q9LPR3 F15H18.4 [Arabidopsis thaliana]
Length = 1702
Score = 459 bits (1180), Expect = e-128
Identities = 224/347 (64%), Positives = 270/347 (77%), Gaps = 6/347 (1%)
Query: 5 KQNSNTFCNQIPNFLSSFIDTFVDFSVSGGLFLPPPPSSPPPI-PTRLPSPSRLIAIGDL 63
++N + C IP +SSF+DTFVD+SVSG +FLP PSS I TR P RL+AIGDL
Sbjct: 4 RENPSGICKSIPKLISSFVDTFVDYSVSG-IFLPQDPSSQNEILQTRFEKPERLVAIGDL 62
Query: 64 HGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEKLKRQAAIH 123
HGDL+KS+EA IAGLIDSS +TGGS VVQ+GDVLDRGG+E+KILY LEKLKR+A
Sbjct: 63 HGDLEKSREAFKIAGLIDSSDRWTGGSTMVVQVGDVLDRGGEELKILYFLEKLKREAERA 122
Query: 124 GGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLE 183
GG +TMNGNHEIMN EGDFR+ TK G+EEF++W +W+ GNKMK LC GL++ DP E
Sbjct: 123 GGKILTMNGNHEIMNIEGDFRYVTKKGLEEFQIWADWYCLGNKMKTLCSGLDKP-KDPYE 181
Query: 184 NVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVD 243
+ ++F +R + +G RAR+AALRP+GPI+KRF T+N TV VVGDS+FVHGGLL EH++
Sbjct: 182 GIPMSFPRMRADCFEGIRARIAALRPDGPIAKRFLTKNQTVAVVGDSVFVHGGLLAEHIE 241
Query: 244 YGLEKINGEVSDWYKGLFGNRFSPPYCRGRNALVWLRKFSD---GNCDCSSLEHVLSTIP 300
YGLE+IN EV W G G R++P YCRG N++VWLRKFS+ CDC++LEH LSTIP
Sbjct: 242 YGLERINEEVRGWINGFKGGRYAPAYCRGGNSVVWLRKFSEEMAHKCDCAALEHALSTIP 301
Query: 301 GVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEIDR 347
GVKRMIMGHTIQ GINGVC +KAIRIDVGMSKGC GLPEVLEI R
Sbjct: 302 GVKRMIMGHTIQDAGINGVCNDKAIRIDVGMSKGCADGLPEVLEIRR 348
>UniRef100_Q8L774 Hypothetical protein At1g07010 [Arabidopsis thaliana]
Length = 389
Score = 177 bits (448), Expect = 6e-43
Identities = 121/329 (36%), Positives = 169/329 (50%), Gaps = 21/329 (6%)
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN--YTGGSATVVQIGDVLDRGG 104
PT + +P+R ++A+GDLHGDL K+++AL +AG++ S G + G +VQ+GD+LDRG
Sbjct: 49 PTFVSAPARRIVAVGDLHGDLGKARDALQLAGVLSSDGRDQWVGQDTVLVQVGDILDRGD 108
Query: 105 DEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEE---FKVWLEWF 161
DEI IL LL L QA +GG +NGNHE MN EGDFR+ +E F +LE +
Sbjct: 109 DEIAILSLLRSLDDQAKANGGAVFQVNGNHETMNVEGDFRYVDARAFDECTDFLDYLEDY 168
Query: 162 RQG--NKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFT 219
Q +N + D + + + G AR LRP G ++
Sbjct: 169 AQDWDKAFRNWIFESRQWKEDRRSSQTYWDQWNVVKRQKGVIARSVLLRPGGRLACELSR 228
Query: 220 QNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGLFGNRFSP--PY--CRGRNA 275
V +L V + +F HGGLL HV YG+E+IN EVS W + SP P+ RG ++
Sbjct: 229 HGV-ILRVNNWLFCHGGLLPHHVAYGIERINREVSTWMRSPTNYEDSPQMPFIATRGYDS 287
Query: 276 LVWLRKFSDGNCDCSSLE--------HVLSTIPGVKRMIMGHTIQKEGINGVCENKAIRI 327
+VW R +S + + H G K M++GHT Q G+N R+
Sbjct: 288 VVWSRLYSRETSELEDYQIEQVNKILHDTLEAVGAKAMVVGHTPQLSGVNCEYGCGIWRV 347
Query: 328 DVGMSKGCGGGLPEVLEIDRYGVRILTSN 356
DVGMS G PEVLEI R++ SN
Sbjct: 348 DVGMSSGVLDSRPEVLEIRGDKARVIRSN 376
>UniRef100_Q8RY10 At1g07010/F10K1_19 [Arabidopsis thaliana]
Length = 389
Score = 173 bits (439), Expect = 7e-42
Identities = 117/333 (35%), Positives = 171/333 (51%), Gaps = 29/333 (8%)
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN--YTGGSATVVQIGDVLDRGG 104
PT + +P+R ++A+GDLHGDL K+++AL +AG++ S G + G +VQ+GD+LDRG
Sbjct: 49 PTFVSAPARRIVAVGDLHGDLGKARDALQLAGVLSSDGRDQWVGQDTVLVQVGDILDRGD 108
Query: 105 DEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQG 164
+EI IL LL L QA +GG +NGNHE MN EGDFR+ +E +L++
Sbjct: 109 EEIAILSLLRSLDDQAKANGGAVFQVNGNHETMNVEGDFRYVDARAFDECTDFLDYLE-- 166
Query: 165 NKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFR---------ARVAALRPNGPISK 215
+ ++ K + + + R + + D + AR LRP G ++
Sbjct: 167 DYAQDWDKAFRNWIFESRQ--WKEDRRSSQTYWDQWNVVKRQKEVIARSVLLRPGGRLAC 224
Query: 216 RFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGLFGNRFSP--PY--CR 271
V +L V + +F HGGLL HV YG+E+IN EVS W + SP P+ R
Sbjct: 225 ELSRHGV-ILRVNNWLFCHGGLLPHHVAYGIERINREVSTWMRSPTNYEDSPQMPFIATR 283
Query: 272 GRNALVWLRKFSDGNCDCSSLE--------HVLSTIPGVKRMIMGHTIQKEGINGVCENK 323
G +++VW R +S + + H G K M++GHT Q G+N
Sbjct: 284 GYDSVVWSRLYSRETSELEDYQIEQVNKILHDTLEAVGAKAMVVGHTPQLSGVNCEYGCG 343
Query: 324 AIRIDVGMSKGCGGGLPEVLEIDRYGVRILTSN 356
R+DVGMS G PEVLEI R++ SN
Sbjct: 344 IWRVDVGMSSGVLDSRPEVLEIRGDKARVIRSN 376
>UniRef100_Q9LMJ5 F10K1.27 protein [Arabidopsis thaliana]
Length = 415
Score = 171 bits (433), Expect = 3e-41
Identities = 122/356 (34%), Positives = 169/356 (47%), Gaps = 49/356 (13%)
Query: 48 PTRLPSPSRLIA----------IGDLHGDLKKSKEALSIAGLIDSSGN--YTGGSATVVQ 95
PT + +P+R I +GDLHGDL K+++AL +AG++ S G + G +VQ
Sbjct: 49 PTFVSAPARRIVAGFGFGYALTVGDLHGDLGKARDALQLAGVLSSDGRDQWVGQDTVLVQ 108
Query: 96 IGDVLDRGGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEF- 154
+GD+LDRG DEI IL LL L QA +GG +NGNHE MN EGDFR+ +E
Sbjct: 109 VGDILDRGDDEIAILSLLRSLDDQAKANGGAVFQVNGNHETMNVEGDFRYVDARAFDECT 168
Query: 155 --------------KVWLEWFRQGNKMKNLCKGLEETVVDPLENVHVAFRGVREEF---- 196
K + W + + K + +T D V + V ++
Sbjct: 169 DFLDYLEDYAQDWDKAFRNWIFESRQWKEDRRS-SQTYWDQWNVVKFLYSLVNQKSVGKP 227
Query: 197 ----HDGFRARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGE 252
G AR LRP G ++ V +L V + +F HGGLL HV YG+E+IN E
Sbjct: 228 LGQRQKGVIARSVLLRPGGRLACELSRHGV-ILRVNNWLFCHGGLLPHHVAYGIERINRE 286
Query: 253 VSDWYKGLFGNRFSP--PY--CRGRNALVWLRKFSDGNCDCSSLE--------HVLSTIP 300
VS W + SP P+ RG +++VW R +S + + H
Sbjct: 287 VSTWMRSPTNYEDSPQMPFIATRGYDSVVWSRLYSRETSELEDYQIEQVNKILHDTLEAV 346
Query: 301 GVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGCGGGLPEVLEIDRYGVRILTSN 356
G K M++GHT Q G+N R+DVGMS G PEVLEI R++ SN
Sbjct: 347 GAKAMVVGHTPQLSGVNCEYGCGIWRVDVGMSSGVLDSRPEVLEIRGDKARVIRSN 402
>UniRef100_Q8L676 Hypothetical protein OSJNBa0011L09.21 [Oryza sativa]
Length = 377
Score = 166 bits (419), Expect = 1e-39
Identities = 106/295 (35%), Positives = 160/295 (53%), Gaps = 28/295 (9%)
Query: 48 PTRLPSPSR-LIAIGDLHGDLKKSKEALSIAGLIDSSGN---YTGGSATVVQIGDVLDRG 103
PT + +P R ++A+GDLHGDL +++ AL +AGL+ S + +TGG +VQ+GD+LDRG
Sbjct: 78 PTFVTAPGRRIVAVGDLHGDLNQTRAALVMAGLLSSESDGHVWTGGQTVLVQVGDILDRG 137
Query: 104 GDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQ 163
DEI IL LL L QA GG +NGNHE +N EGD+R+ +E ++E+ +
Sbjct: 138 EDEIAILSLLSSLNMQAKSQGGAVFQVNGNHETINVEGDYRYVDPGAFDECIRFMEYLDE 197
Query: 164 --GN------KMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISK 215
GN N+C+ +E + V+++ G AR + + GP++
Sbjct: 198 CDGNWDDAFLNWVNVCERWKEEYPMSPNGDWRPWNFVKKQ--KGIAARSSLFKRGGPLAC 255
Query: 216 RFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGLFG---NRFSPPY--C 270
++ VL V D IF HGGLL HV+YG+E++N EVS W K G + P+
Sbjct: 256 E-LARHPVVLSVNDWIFCHGGLLPHHVEYGIERMNREVSVWMKSSSGDSDDELDIPFIAT 314
Query: 271 RGRNALVWLRKFSDGNCDCSSLEHVLSTI--------PGVKRMIMGHTIQKEGIN 317
RG +++VW R +S G + + LS++ G K M++GHT Q G+N
Sbjct: 315 RGYDSVVWSRLYSQGPTEMTRHSWKLSSVVAERTLKSVGAKGMVVGHTPQTRGVN 369
>UniRef100_UPI00002D058A UPI00002D058A UniRef100 entry
Length = 182
Score = 141 bits (355), Expect = 4e-32
Identities = 72/153 (47%), Positives = 93/153 (60%), Gaps = 4/153 (2%)
Query: 201 RARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKGL 260
R R AL P P++ RF V+ VG ++FVHGG+L EHV YGL+ +N E+S W K
Sbjct: 10 RDRWRALAPGAPLTLRFLAHQPVVVAVGSTLFVHGGILPEHVQYGLDSLNAEISTWMKMG 69
Query: 261 FGNRFSPPYCRGRNALVWLRKFS---DGNCDCSSLEHVLSTIPGVKRMIMGHTIQ-KEGI 316
P +GR++LVW R +S + CDC L+ L IPGV+R+++GHTIQ GI
Sbjct: 70 KIKNAPPMSVQGRDSLVWARDYSHVQEHKCDCDLLKETLKMIPGVERIVVGHTIQHPHGI 129
Query: 317 NGVCENKAIRIDVGMSKGCGGGLPEVLEIDRYG 349
C+ K IR+DVGMSKGC PE LEI R G
Sbjct: 130 TSACDGKVIRVDVGMSKGCVDAKPEALEILRDG 162
>UniRef100_UPI000042F2AE UPI000042F2AE UniRef100 entry
Length = 385
Score = 117 bits (292), Expect = 7e-25
Identities = 93/308 (30%), Positives = 141/308 (45%), Gaps = 80/308 (25%)
Query: 56 RLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEK 115
R++ IGD+HGDLK +L ++G+I+S+ ++ + ++Q+GDV+DRG ++I L K
Sbjct: 87 RVLVIGDIHGDLKSLITSLFLSGVINSNLDWIAKNTLLIQLGDVVDRGSHALQIYKLFNK 146
Query: 116 LKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVW-------LEWFRQGNKMK 168
LK QA G F+ + GNHE+MN G + T E+F+ + EW ++G K
Sbjct: 147 LKSQAPSLGSKFVGLLGNHEVMNLCGQLHYVTD---EDFQTYGGRDNRTFEWSKEGFVGK 203
Query: 169 NLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVVG 228
L R A+R V
Sbjct: 204 YL------------------------------RTMKLAIR------------------VN 215
Query: 229 DSIFVHGGLLKEHVDYGLEKIN--------GEVSDWYKGLFGNRFSPPYCRGRNALVWLR 280
DS++VH GLL ++ GL+K++ G+ D+Y LF P +W R
Sbjct: 216 DSLYVHAGLLPKYAKLGLDKLDKLSNDLLEGDFCDFYSSLFFVEDGP---------LWTR 266
Query: 281 KFSDGNCD--CSSLEHVLSTIPGVKRMIMGHTIQKEG-INGVCENKAIRIDVGMSKGCGG 337
S G + C ++ L I G+ RM++GHTIQ + IN C+NK I D G S+
Sbjct: 267 DISLGEEEKACKLVDETLQ-ILGLSRMVVGHTIQHDNRINIKCDNKLILADTGFSEAI-Y 324
Query: 338 GLPEVLEI 345
G P +LEI
Sbjct: 325 GKPCMLEI 332
>UniRef100_UPI0000452C3B UPI0000452C3B UniRef100 entry
Length = 385
Score = 115 bits (288), Expect = 2e-24
Identities = 91/308 (29%), Positives = 141/308 (45%), Gaps = 80/308 (25%)
Query: 56 RLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGGDEIKILYLLEK 115
R++ IGD+HGDLK +L ++G+I+S+ ++ + ++Q+GDV+DRG ++I L K
Sbjct: 87 RVLVIGDIHGDLKSLITSLFLSGVINSNLDWIAKNTLLIQLGDVVDRGSHALQIYKLFNK 146
Query: 116 LKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVW-------LEWFRQGNKMK 168
LK QA G F+ + GNHE+MN G + T E+F+ + EW ++G K
Sbjct: 147 LKSQAPSLGSKFVGLLGNHEVMNLCGQLHYVTD---EDFQTYGGRDNRTFEWSKEGFVGK 203
Query: 169 NLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVVG 228
L R A+R V
Sbjct: 204 YL------------------------------RTMKLAIR------------------VN 215
Query: 229 DSIFVHGGLLKEHVDYGLEKIN--------GEVSDWYKGLFGNRFSPPYCRGRNALVWLR 280
DS++VH GLL ++ GL++++ G+ D+Y LF P +W R
Sbjct: 216 DSLYVHAGLLPKYAKLGLDRLDKLSNDLLEGDFCDFYSSLFFVEDGP---------LWTR 266
Query: 281 KFSDGNCD--CSSLEHVLSTIPGVKRMIMGHTIQKEG-INGVCENKAIRIDVGMSKGCGG 337
S G + C ++ L I G+ RM++GHTIQ + IN C+NK + D G S+
Sbjct: 267 DISLGEEEKACKLVDETLQ-ILGLSRMVVGHTIQHDNRINIKCDNKLVLADTGFSEAI-Y 324
Query: 338 GLPEVLEI 345
G P +LEI
Sbjct: 325 GKPCMLEI 332
>UniRef100_UPI0000280CED UPI0000280CED UniRef100 entry
Length = 320
Score = 107 bits (267), Expect = 6e-22
Identities = 72/243 (29%), Positives = 115/243 (46%), Gaps = 59/243 (24%)
Query: 50 RLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSG-------------NYTGGSATVVQI 96
+LP +++ AIGDLHGDL+ ++L +AG+I S + GG+ VVQ+
Sbjct: 75 KLPEGAKIAAIGDLHGDLEVCIKSLKLAGVISLSTPHYFETIQDANNIEWIGGNTHVVQV 134
Query: 97 GDVLDR-------------------GGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIM 137
GD +DR G ++KI+ LL++L +A GG I++ GNHEIM
Sbjct: 135 GDQIDRCRPNNWYRDICADDSTYQDEGSDLKIMELLDRLNEKAKYQGGAIISILGNHEIM 194
Query: 138 NAEGDFRFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLENVHVAFRGVREEFH 197
N GDFR+ + EEF + + + N F
Sbjct: 195 NCVGDFRYVSPKEFEEFGEFYKIKKTNNP--------------------------NRVFP 228
Query: 198 DGFRARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLL-KEHVDYGLEKINGEVSDW 256
G++ R A P G I+++F +Q +++ +G +FVHGG+ K +Y L+++N +S W
Sbjct: 229 YGYKERKQAFSPGGIIARKFASQRHSIVQIGGWVFVHGGITPKLSNEYTLDEMNEGMSKW 288
Query: 257 YKG 259
G
Sbjct: 289 LLG 291
>UniRef100_UPI000034C07C UPI000034C07C UniRef100 entry
Length = 180
Score = 101 bits (251), Expect = 4e-20
Identities = 45/100 (45%), Positives = 68/100 (68%)
Query: 44 PPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRG 103
P P+ + RL+A+GD+HGD ++ +AL ++ +DS G + GG+ +VQ+GD+LDRG
Sbjct: 65 PKPVTNLDLNGRRLVAVGDIHGDFAQAMKALELSKCMDSEGKWIGGTTVLVQVGDILDRG 124
Query: 104 GDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDF 143
+E+ I+ +KL R+A GG+ + MNGNHEIMN GDF
Sbjct: 125 DNELAIMRKFQKLAREAKEAGGDVVVMNGNHEIMNVMGDF 164
>UniRef100_UPI00003C1C90 UPI00003C1C90 UniRef100 entry
Length = 614
Score = 100 bits (249), Expect = 7e-20
Identities = 88/302 (29%), Positives = 135/302 (44%), Gaps = 50/302 (16%)
Query: 56 RLIAIGDLHGDLKKSKEALSIAGLI------DSSGNYTGGSATVVQIGDVLDRGGDEIKI 109
R +A+ DLHGDL + LS+A ++ D + + GG T+V GD++DRG D I +
Sbjct: 291 RTVAVADLHGDLDHALNVLSMASIVSRTRSADHAYTWIGGHDTLVSTGDIVDRGDDTIAL 350
Query: 110 LYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNKMKN 169
L L++QA + GG GNHE+MNA GD+R+ TK VE F G + +
Sbjct: 351 YRLFVSLRQQARLAGGEVKNCLGNHEVMNAIGDWRYVTKADVESFG--------GVQARR 402
Query: 170 LCKGLEETVVDP-LENVHVAFR-GVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLVV 227
+ + L N +V + E H AL PN + V+
Sbjct: 403 HAMSDQGWIGQEWLHNYNVTHTISLLPETHP-------ALPPN------YTPPRVS---- 445
Query: 228 GDSIFVHGGLLKEHVDYGLEKINGEV-SDWYKGLFG--NRFSPPYCRG-------RNALV 277
FVHGG+ ++ G++ IN S KGL N + PP + +
Sbjct: 446 ----FVHGGITPQYAALGIDFINTVGHSLLLKGLSSQPNSWLPPNTTSDEQALWSEHGPL 501
Query: 278 WLRKFSDGNCD--CSSLEHVLSTIPGVKRMIMGHTIQKEGINGVCENKAIRIDVGMSKGC 335
W R ++ C++ E ++ V +++MGHT +G C N + ID G+S+
Sbjct: 502 WYRGYATDRLSHACANAEKATKSL-AVNQLVMGHTPHFDGFVTRCNNTILLIDTGISRAY 560
Query: 336 GG 337
GG
Sbjct: 561 GG 562
>UniRef100_O74480 SPCC1840.07c protein [Schizosaccharomyces pombe]
Length = 332
Score = 100 bits (248), Expect = 9e-20
Identities = 86/313 (27%), Positives = 131/313 (41%), Gaps = 56/313 (17%)
Query: 36 FLPPPPSSPPPIPTRLPSPSRLIAIGDLHGDLKKSKEALSIAGLIDS--SGNYTGGSATV 93
F+ P+ + ++ P+ + AIGD+HGD + + LS AG++ +T G+AT+
Sbjct: 38 FIKVFPNFKGNVEAKIKYPTVVYAIGDIHGDFPNALDVLSAAGVVSPVFPHEWTAGNATL 97
Query: 94 VQIGDVLDRGGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRF------AT 147
VQ GDV+DRG D K+ L +QA HGG + + GNHE MNA+GD+R+ A+
Sbjct: 98 VQTGDVVDRGPDTRKLFRWFNDLHKQAEKHGGRVVRLLGNHEFMNAKGDWRYVHPGDKAS 157
Query: 148 KNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAAL 207
E ++W G L T D H G
Sbjct: 158 YPEPSEENRIIDWGHSGEIGNLLLSEYNVTYKDNTTGSHFMHAG---------------- 201
Query: 208 RPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLE---KINGEVSDWYKGLFGNR 264
+S + + TV +G + H + +E + LE I G + WY+G
Sbjct: 202 -----LSPEWAYREETVNELGKELLSH-FMSREKIPEYLEDFWAIEGPM--WYRG----- 248
Query: 265 FSPPYCRGRNALVWLRKFSDGNCDCSSLEHVLSTIPGVKRMIMGHTIQKEGINGVCENKA 324
L + S+ C +V T+ V R++MGHT Q GI CE +
Sbjct: 249 --------------LAQLSEEEA-CEVALNVTKTL-NVNRLVMGHTPQFHGIVSRCEGRI 292
Query: 325 IRIDVGMSKGCGG 337
+ ID G+ G
Sbjct: 293 LLIDTGLCSAYAG 305
>UniRef100_UPI00002F6C5D UPI00002F6C5D UniRef100 entry
Length = 360
Score = 98.6 bits (244), Expect = 3e-19
Identities = 90/333 (27%), Positives = 147/333 (44%), Gaps = 75/333 (22%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGN--------YTGGSATVVQIGDVLDR- 102
P +LIA+GD+HGDL+ + AL +A +I + N + GGS ++Q+GD +DR
Sbjct: 3 PPVKKLIAMGDVHGDLRATIIALKLAEVIPQTANEQNINSMHWCGGSTWLIQLGDQIDRC 62
Query: 103 -------------------GGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDF 143
G+ +KI+ L +L +A GG + + GNHE+MN + DF
Sbjct: 63 RPDELEKNCIKDFSDVFEDEGNNMKIIQLFLRLDDEARKEGGRVLGLLGNHELMNIDKDF 122
Query: 144 RFATKNGVEEFKVWLEWFRQGNKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRAR 203
R+ + +EF LE+ Q ++ K + + G+ R
Sbjct: 123 RYVSP---QEF---LEFVPQKDRDSKYTK---------------------DGYPLGYYHR 155
Query: 204 VAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVD-YGLEKINGEVSDWYK---- 258
A + G IS + + +++V+G IFVHGGL ++ V+ Y + +IN VS W +
Sbjct: 156 TKAFQRGGNISTMYGLKKKSIMVIGGFIFVHGGLSRDLVNKYTIHEINNVVSKWLQKKSN 215
Query: 259 ----GLFGNRF------SPPYCR--GRNALVWLRKFSDGNCDCSSLEHVLSTIPGVKRMI 306
+F F SP +CR + V N L + ++R++
Sbjct: 216 DTEDNIFDEIFRDDDDMSPFWCRLYSEDDGVGENTTKGFNQLLKILNQRNKKLQPIERIV 275
Query: 307 MGHTIQKEG---INGVCENKAIRIDVGMSKGCG 336
+ HT Q +N + + RIDVGMS+ G
Sbjct: 276 LAHTPQFMNDMYMNSLYNERLWRIDVGMSRAFG 308
>UniRef100_UPI00002B5187 UPI00002B5187 UniRef100 entry
Length = 224
Score = 97.8 bits (242), Expect = 5e-19
Identities = 66/251 (26%), Positives = 120/251 (47%), Gaps = 56/251 (22%)
Query: 56 RLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGDVLDRGG---------DE 106
R++AIGD+HGD + ++L + +I+ + N+ G + ++Q+GD+LDRGG E
Sbjct: 16 RIVAIGDVHGDYVATIKSLQLVNIINKNLNWIGKNTILIQMGDILDRGGRDVTYGDEDSE 75
Query: 107 IKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFRQGNK 166
IKI+YL ++LK+QA + GN I + GNHE++N +G + + +K G+++F G K
Sbjct: 76 IKIIYLFKRLKKQAIKNKGNVICIIGNHEMLNLQGVYDYCSKLGIKKF---------GTK 126
Query: 167 MKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNVTVLV 226
K R++F+ +P ++ +
Sbjct: 127 KK------------------------RKKFY----------KPGNKMAISMTKLFKAIYK 152
Query: 227 VGDSIFVHGGLLKE-HVDYGLEKINGEVSDWYKGLFGNRFSPPYCR---GRNALVWLRKF 282
+G IFVHGG+ Y ++ IN + ++ G S + N+++W R +
Sbjct: 153 IGPWIFVHGGIRPYLSKKYTIDYINKTMHEYLSGNINLEDSKEFQELFLDENSILWYRGY 212
Query: 283 SDGNCDCSSLE 293
++G +C L+
Sbjct: 213 AEGTINCRQLK 223
>UniRef100_UPI000042F3C1 UPI000042F3C1 UniRef100 entry
Length = 385
Score = 94.7 bits (234), Expect = 4e-18
Identities = 46/116 (39%), Positives = 71/116 (60%), Gaps = 4/116 (3%)
Query: 43 SPPPIPTRLPSPS----RLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTGGSATVVQIGD 98
S P +P+R + RL+A+GDLHGD+ +K+ L +A +ID + + +VQ GD
Sbjct: 32 SVPNVPSRGAGEAAYRQRLVAVGDLHGDIDNAKKTLQMARIIDDDSKWVASTDILVQTGD 91
Query: 99 VLDRGGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEF 154
++DRG I L++ L+ QAA GG +++ GNHE+MNA GD+R+ TK + F
Sbjct: 92 IVDRGAYADDIYRLMQSLRGQAASQGGKVVSILGNHEVMNAIGDWRYVTKGDIARF 147
>UniRef100_UPI000030DC8C UPI000030DC8C UniRef100 entry
Length = 247
Score = 88.2 bits (217), Expect = 4e-16
Identities = 56/200 (28%), Positives = 89/200 (44%), Gaps = 52/200 (26%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSGNYTG--GSATVVQIGDVLDRGG----- 104
P R+I IGDLHGD +K+ LID+S N+ + VVQ+GD LD GG
Sbjct: 13 PKKDRIIVIGDLHGDYQKTISLFKSLKLIDNSLNWVAFPKNTFVVQLGDQLDGGGRGEGD 72
Query: 105 --DEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDFRFATKNGVEEFKVWLEWFR 162
E+K++ +E + ++A + GG +++ GNHE+MN GDFR+A+++ +EE
Sbjct: 73 TRGELKLINFMEDINQKAQVVGGAVLSLIGNHEVMNLIGDFRYASEHDIEEV-------- 124
Query: 163 QGNKMKNLCKGLEETVVDPLENVHVAFRGVREEFHDGFRARVAALRPNGPISKRFFTQNV 222
G R +P G + +
Sbjct: 125 -----------------------------------GGLNVRKKLFKPGGELFNKLSCTRN 149
Query: 223 TVLVVGDSIFVHGGLLKEHV 242
V+ +G +FVH G+L +H+
Sbjct: 150 VVVKIGSWVFVHAGVLPKHI 169
>UniRef100_UPI00002B8E83 UPI00002B8E83 UniRef100 entry
Length = 309
Score = 84.3 bits (207), Expect = 5e-15
Identities = 59/165 (35%), Positives = 85/165 (50%), Gaps = 11/165 (6%)
Query: 201 RARVAALRPNGPISKRFFTQNVTVLVVGDSIFVHGGLLKEHVDYGLEKINGEVSDWYKG- 259
+ RV A RP G +KR V + V+GD++ VHGGL ++HV YG+E++N E++ W G
Sbjct: 15 KQRVQAFRPGGFEAKRLSKMPVAI-VIGDTLLVHGGLREKHVKYGVERMNKEMAAWLAGP 73
Query: 260 -LFGNRFSPPYCRGRNALVWLRKFSDGNCDC---SSLEHVLSTIPGVKRMIMGHTIQKEG 315
F P ++ +W R +S + LE V + VKRM++GHT Q G
Sbjct: 74 PKFRGADKPEIIDESDSPIWARLYSTPTPKSFAETELEEVCKALK-VKRMVVGHTPQLRG 132
Query: 316 INGVCEN---KAIRIDVGMSKGCGGGLPEVLEIDRYG-VRILTSN 356
IN + R D GMS G G E LE+ R G +LT++
Sbjct: 133 INAFVSEGNYEVWRTDTGMSSGMMSGPLECLEVLRDGTTHVLTTD 177
>UniRef100_UPI00002C3AD7 UPI00002C3AD7 UniRef100 entry
Length = 223
Score = 77.0 bits (188), Expect = 8e-13
Identities = 47/131 (35%), Positives = 69/131 (51%), Gaps = 28/131 (21%)
Query: 52 PSPSRLIAIGDLHGDLKKSKEALSIAGLIDSSG--------NYTGGSATVVQIGDVLDR- 102
P RL+AIGDLHGDL+ + AL +A +I + ++TGGS+ VVQ+GD +DR
Sbjct: 48 PPVKRLVAIGDLHGDLRATVLALKLAEVISQNSTPENVNMIHWTGGSSWVVQLGDQIDRC 107
Query: 103 -------------------GGDEIKILYLLEKLKRQAAIHGGNFITMNGNHEIMNAEGDF 143
G+ + I+ LL +L +A GG F+ + GNHE+MN + DF
Sbjct: 108 RPDELEKNCIKDFSDVFEDEGNNMTIIKLLLRLDDEARKEGGRFLGLLGNHELMNIDKDF 167
Query: 144 RFATKNGVEEF 154
R+ + EF
Sbjct: 168 RYVSPQEFLEF 178
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.141 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 733,171,067
Number of Sequences: 2790947
Number of extensions: 34018987
Number of successful extensions: 175746
Number of sequences better than 10.0: 222
Number of HSP's better than 10.0 without gapping: 66
Number of HSP's successfully gapped in prelim test: 168
Number of HSP's that attempted gapping in prelim test: 175125
Number of HSP's gapped (non-prelim): 448
length of query: 394
length of database: 848,049,833
effective HSP length: 129
effective length of query: 265
effective length of database: 488,017,670
effective search space: 129324682550
effective search space used: 129324682550
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC146752.2


