

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146704.8 - phase: 1 /pseudo
(946 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
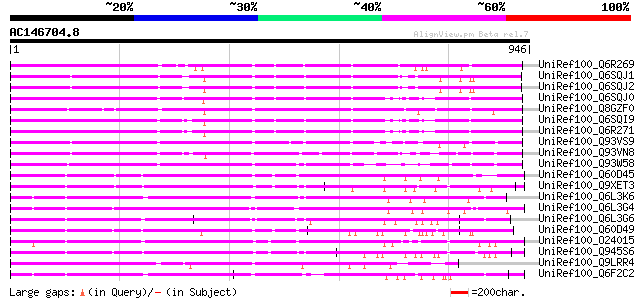
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q6R269 Disease resistance protein [Glycine max] 603 e-171
UniRef100_Q6SQJ1 NBS-LRR type disease resistance protein RPG1-B ... 565 e-159
UniRef100_Q6SQJ2 NBS-LRR type disease resistance protein RPG1-B ... 564 e-159
UniRef100_Q6SQJ0 NBS-LRR type disease resistance protein Hom-F [... 563 e-159
UniRef100_Q8GZF0 Resistance protein KR4 [Glycine max] 563 e-158
UniRef100_Q6SQI9 NBS-LRR type disease resistance protein Hom-B [... 561 e-158
UniRef100_Q6R271 Disease resistance protein [Glycine max] 561 e-158
UniRef100_Q93VS9 NBS-LRR resistance-like protein B8 [Phaseolus v... 543 e-153
UniRef100_Q93VN8 NBS-LRR resistance-like protein B11 [Phaseolus ... 538 e-151
UniRef100_Q93W58 NBS-LRR resistance-like protein J71 [Phaseolus ... 519 e-145
UniRef100_Q60D45 Putative disease resistance protein I2 [Solanum... 516 e-144
UniRef100_Q9XET3 Disease resistance protein I2 [Lycopersicon esc... 515 e-144
UniRef100_Q6L3K6 Putative resistance complex protein I2C-2 [Sola... 514 e-144
UniRef100_Q6L3G4 Putative plant disease resistant protein [Solan... 497 e-139
UniRef100_Q6L3G6 Putative plant disease resistant protein [Solan... 496 e-138
UniRef100_Q60D49 Putative disease resistance protein I2C-5 [Sola... 496 e-138
UniRef100_O24015 Resistance complex protein I2C-1 [Lycopersicon ... 482 e-134
UniRef100_Q945S6 I2C-5 [Lycopersicon pimpinellifolium] 477 e-133
UniRef100_Q9LRR4 Putative disease resistance RPP13-like protein ... 464 e-129
UniRef100_Q6F2C2 Putative disease resistance protein [Solanum de... 461 e-128
>UniRef100_Q6R269 Disease resistance protein [Glycine max]
Length = 1189
Score = 603 bits (1556), Expect = e-171
Identities = 388/994 (39%), Positives = 563/994 (56%), Gaps = 88/994 (8%)
Query: 1 YNDNKIKE-HFELKAWVYVSESFDVVGLTKAILKSFNSSAD--GEDLNLLQHQLQYMLMG 57
YN+ +I+E F++K W+ VS+ FDV+ L+K IL S D G+DL ++ +L+ L G
Sbjct: 212 YNNPRIEEAKFDIKVWICVSDDFDVLMLSKTILNKITKSKDDSGDDLEMVHGRLKEKLSG 271
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
KYL VLDD+WN D ++W+ L P +G+ GSKI+VTTR +VA + ++S ++ +L+QL
Sbjct: 272 NKYLFVLDDVWNEDRDQWKALQTPLKYGAKGSKILVTTRSNKVA-STMQSNKVHELKQLQ 330
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
+ + W +F HAFQ L+ +G KI+EKC GLPLA++++G LL K S +W
Sbjct: 331 EDHSWQVFAQHAFQDDYPKLNAELKEIGIKIIEKCQGLPLALETVGCLLHKKPSISQWEG 390
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
+L++ +W L+K + + L LSY++LPS+LKRCF+YC++FPK H+F KD LI LW+AE
Sbjct: 391 VLKSKIWELTKEESKIIPALLLSYYHLPSHLKRCFAYCALFPKDHEFYKDSLIQLWVAEN 450
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFS 297
++C + S+EE G + F DL+S SFFQ+S E + + MHDL+NDL K V G+
Sbjct: 451 FVQCSQQSNSQEEIGEQYFNDLLSRSFFQRSSIE-----KCFFMHDLLNDLAKYVCGDIC 505
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSVDKLLELTCEGLHS----LILEGTRAMLISN--- 350
++E + +S+ + RH F + +D+ + H+ + TR +L++N
Sbjct: 506 FRLEVDK-PKSISKVRHFSFVTE---IDQYFDGYGSLYHAQRLRTFMPMTRPLLLTNWGG 561
Query: 351 -NVQQDLFSRLNFLRMLSFRGCGLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHNL 409
+ +L S+ FLR+LS C L E+ D + NL LR LDLSYT+I+ LPD++C L NL
Sbjct: 562 RKLVDELCSKFKFLRILSLFRCDLKEMPDSVGNLNHLRSLDLSYTFIKKLPDSMCFLCNL 621
Query: 410 QTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVE 469
Q L L C L ELPSN KL NLR L+ ++ MP H GKL NLQ LS F V
Sbjct: 622 QVLKLNYCVHLEELPSNLHKLTNLRCLEFMCTK----VRKMPMHMGKLKNLQVLSPFYVG 677
Query: 470 E--QNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREE 527
+ N S +++L +LN LHG++ IE L N+ + D+ +LK+ +L +L ++++ R
Sbjct: 678 KGIDNCS-IQQLGELN-LHGSLSIEELQNIVNPLDALAABLKNKTHLLDLRLEWNEDR-N 734
Query: 528 MDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSL 587
+D+S+ E VLE LQP+R+L++L+I Y G FP+W+ L N+VSL L C
Sbjct: 735 LDDSIKER--QVLENLQPSRHLEKLSIRNYGGTQFPSWLSDNSLCNVVSLTLMNCKYFLC 792
Query: 588 LPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC--- 644
LPPLG LP LK LSI DGI I +F+ SSS + F SLE LKF M WEEW C
Sbjct: 793 LPPLGLLPILKELSIEGLDGIVSINADFFGSSSCS--FTSLESLKFSDMKEWEEWECKGV 850
Query: 645 LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDR 704
FP L+ L I+ CPKLK LP+ L L L I+ C+ L S + +I L + C +
Sbjct: 851 TGAFPRLQRLSIKRCPKLKGHLPEQLCHLNGLKISGCEQLVPSALSAPDIHQLYLGDCGK 910
Query: 705 ILVNELPTSLKKLFILENRYTEFSVEQIFVNSTI-------------------------L 739
+ ++ PT+LK+L I + +EQI N + L
Sbjct: 911 LQIDH-PTTLKELTITGHNMEAALLEQIGRNYSCSNKNIPMHSCYDFLVWLLINGGCDSL 969
Query: 740 EVLELDLNGSLK------CPTLDLC----CYNSLGELSITRWCS--SSLSFSLH-LFTNL 786
+ LD+ LK CP L +N L +LS+ R C SL +H L +L
Sbjct: 970 TTIHLDIFPKLKELYICQCPNLQRISQGQAHNHLQDLSM-RECPQLESLPEGMHVLLPSL 1028
Query: 787 YSLWFVDCPNLDSFPEGGLPCNLLSLTI-TNCPKLI-----ASRQEWGLKSLKYFFVCDD 840
SLW + CP ++ FPEGGLP NL +++ KLI A L+SL V
Sbjct: 1029 DSLWIIHCPKVEMFPEGGLPSNLKVMSLHGGSYKLIYLLKSALGGNHSLESLSIGGV--- 1085
Query: 841 FENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPEEA 900
+VE P E +LP +L L +N C L+ ++ +G HL SL+ L + CP L+ LPEE
Sbjct: 1086 --DVECLPDEGVLPHSLVTLMINKCGDLKRLDYKGLCHLSSLKRLSLWECPRLQCLPEEG 1143
Query: 901 LPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHI 934
LP S+ +L I +CPL+K + ++ GE I HI
Sbjct: 1144 LPKSISTLRILNCPLLKQRCREPEGEDWPKIAHI 1177
>UniRef100_Q6SQJ1 NBS-LRR type disease resistance protein RPG1-B [Glycine max]
Length = 1217
Score = 565 bits (1457), Expect = e-159
Identities = 371/1000 (37%), Positives = 544/1000 (54%), Gaps = 106/1000 (10%)
Query: 1 YNDNKIKE-HFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGK 58
+ND +I+E F++KAWV VS+ FD +T+ IL++ S D DL ++ +L+ L GK
Sbjct: 228 FNDPRIEEARFDVKAWVCVSDDFDAFRVTRTILEAITKSTDDSRDLEMVHGRLKEKLTGK 287
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
++LLVLDD+WN + +WE +L G+ GS+I+ TTR KEVA + L L+QL +
Sbjct: 288 RFLLVLDDVWNENRLKWEAVLKHLGFGAQGSRIIATTRSKEVASTMRSKEHL--LEQLQE 345
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
+CW LF HAFQ ++ P+ + +G KIVEKC GLPLA+K++G LL S EW +I
Sbjct: 346 DHCWKLFAKHAFQDDNIQPNPDCKEIGMKIVEKCKGLPLALKTMGSLLHNKSSVTEWKSI 405
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
L++++W S ++ L LSYH+LPS+LKRCF+YC++FPK ++F K+ LI LWMAE
Sbjct: 406 LQSEIWEFSTERSDIVPALALSYHHLPSHLKRCFAYCALFPKDYEFDKECLIQLWMAEKF 465
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
L+C +S E G + F DL+S FFQQS + +VMHDL+NDL + + G+
Sbjct: 466 LQCSQQGKSPGEVGEQYFNDLLSRCFFQQSSN---TERTDFVMHDLLNDLARFICGDICF 522
Query: 299 QIEDARVERSVERTRHIWFSLQSNSVDKLLELTC-EGLHSLILEGTRAMLISNNVQ---- 353
+++ + + + + TRH L+++ C +G +L + + +
Sbjct: 523 RLDGNQTKGTPKATRHF-----------LIDVKCFDGFGTLCDTKKLRTYMPTSYKYWDC 571
Query: 354 ----QDLFSRLNFLRMLSFRGC-GLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHN 408
+LFS+ N+LR+LS C L E+ D + NLK LR LDLS T IE LP++IC L+N
Sbjct: 572 EMSIHELFSKFNYLRVLSLFDCHDLREVPDSVGNLKYLRSLDLSNTKIEKLPESICSLYN 631
Query: 409 LQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL-SYFI 467
LQ L L GC L ELPSN KL +L L+L ++ +P H GKL LQ L S F
Sbjct: 632 LQILKLNGCRHLKELPSNLHKLTDLHRLELIETG----VRKVPAHLGKLEYLQVLMSSFN 687
Query: 468 VEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREE 527
V + +++L +LN LHG++ I L NV + +D+ V+LK+ +L EL +++D
Sbjct: 688 VGKSREFSIQQLGELN-LHGSLSIRQLQNVENPSDALAVDLKNKTHLVELELEWDSDWNP 746
Query: 528 MDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSL 587
D+S E + V+E LQP+++L++L + Y G FP W+ ++VSL L+ C C
Sbjct: 747 -DDSTKERD--VIENLQPSKHLEKLRMRNYGGTQFPRWLFNNSSCSVVSLTLKNCKYCLC 803
Query: 588 LPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC--- 644
LPPLG LP LK LSI DGI I +F+ SSS + F SL+ L+F M WEEW C
Sbjct: 804 LPPLGLLPSLKELSIKGLDGIVSINADFFGSSSCS--FTSLKSLEFYHMKEWEEWECKGV 861
Query: 645 LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDR 704
FP L+ L I CPKLK LP+ L L L I+ C+ L S + +I L + C
Sbjct: 862 TGAFPRLQRLSIERCPKLKGHLPEQLCHLNSLKISGCEQLVPSALSAPDIHKLYLGDCGE 921
Query: 705 ILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDL-CCYNSL 763
+ ++ T+LK E ++E V + + E E+ N S + + CY+ L
Sbjct: 922 LQIDH-GTTLK----------ELTIEGHNVEAALFE--EIGRNYSCSNNNIPMHSCYDFL 968
Query: 764 GELSITRWCSSSLSFSLHLFT-----------------------NLYSLWFVDCPNLDSF 800
L I C S +F L +FT +L +L +CP L+S
Sbjct: 969 VSLRIKGGCDSLTTFPLDMFTILRELCIWKCPNLRRISQGQAHNHLQTLDIKECPQLESL 1028
Query: 801 PEG--GLPCNLLSLTITNCPK--------LIASRQEWGLKSLKYFFVC---------DDF 841
PEG L +L SL I +CPK L ++ +E GL Y +
Sbjct: 1029 PEGMHVLLPSLDSLCIDDCPKVEMFPEGGLPSNLKEMGLFGGSYKLMSLLKSALGGNHSL 1088
Query: 842 ENV-------ESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLE 894
E + E P+E +LP +L L +N+C L+ ++ +G HL SL+ L + +CP L+
Sbjct: 1089 ERLVIGKVDFECLPEEGVLPHSLVSLQINSCGDLKRLDYKGICHLSSLKELSLEDCPRLQ 1148
Query: 895 RLPEEALPNSLYSLWI-KDCPLIKVKYQKEGGEQRDTICH 933
LPEE LP S+ SLWI DC L+K + ++ GE I H
Sbjct: 1149 CLPEEGLPKSISSLWIWGDCQLLKERCREPEGEDWPKIAH 1188
>UniRef100_Q6SQJ2 NBS-LRR type disease resistance protein RPG1-B [Glycine max]
Length = 1217
Score = 564 bits (1453), Expect = e-159
Identities = 369/1000 (36%), Positives = 544/1000 (53%), Gaps = 106/1000 (10%)
Query: 1 YNDNKIKE-HFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGK 58
+ND +I+E F++KAWV VS+ FD +T+ IL++ S D DL ++ +L+ L GK
Sbjct: 228 FNDPRIEEARFDVKAWVCVSDDFDAFRVTRTILEAITKSTDDSRDLEMVHGRLKEKLTGK 287
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
++LLVLDD+WN + +WE +L G+ GS+I+ TTR KEVA + L L+QL +
Sbjct: 288 RFLLVLDDVWNENRLKWEAVLKHLGFGAQGSRIIATTRSKEVASTMRSKEHL--LEQLQE 345
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
+CW LF HAFQ ++ P+ + +G KIVEKC GLPLA+K++G LL S EW +I
Sbjct: 346 DHCWKLFAKHAFQDDNIQPNPDCKEIGMKIVEKCKGLPLALKTMGSLLHNKSSVTEWKSI 405
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
L++++W S ++ L LSYH+LPS+LKRCF+YC++FPK ++F K+ LI LWMAE
Sbjct: 406 LQSEIWEFSTERSDIVPALALSYHHLPSHLKRCFAYCALFPKDYEFDKECLIQLWMAEKF 465
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
L+C +S E G + F DL+S FFQQS + +VMHDL+NDL + + G+
Sbjct: 466 LQCSQQGKSPGEVGEQYFNDLLSRCFFQQSSN---TERTDFVMHDLLNDLARFICGDICF 522
Query: 299 QIEDARVERSVERTRHIWFSLQSNSVDKLLELTC-EGLHSLILEGTRAMLISNNVQ---- 353
+++ + + + + TRH L+++ C +G +L + + +
Sbjct: 523 RLDGNQTKGTPKATRHF-----------LIDVKCFDGFGTLCDTKKLRTYMPTSYKYWDC 571
Query: 354 ----QDLFSRLNFLRMLSFRGC-GLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHN 408
+LFS+ N+LR+LS C L E+ D + NLK LR LDLS T IE LP++IC L+N
Sbjct: 572 EMSIHELFSKFNYLRVLSLFDCHDLREVPDSVGNLKYLRSLDLSNTKIEKLPESICSLYN 631
Query: 409 LQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL-SYFI 467
LQ L L GC L ELPSN KL +L L+L ++ +P H GKL LQ L S F
Sbjct: 632 LQILKLNGCRHLKELPSNLHKLTDLHRLELIETG----VRKVPAHLGKLEYLQVLMSSFN 687
Query: 468 VEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREE 527
V + +++L +LN LHG++ I L NV + +D+ V+LK+ +L E+ +++D
Sbjct: 688 VGKSREFSIQQLGELN-LHGSLSIRQLQNVENPSDALAVDLKNKTHLVEVELEWDSDWNP 746
Query: 528 MDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSL 587
D+S E + V+E LQP+++L++L + Y G FP W+ ++VSL L+ C C
Sbjct: 747 -DDSTKERD--VIENLQPSKHLEKLRMRNYGGTQFPRWLFNNSSCSVVSLTLKNCKYCLC 803
Query: 588 LPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC--- 644
LPPLG LP LK LSI DGI I +F+ SSS + F SL+ L+F M WEEW C
Sbjct: 804 LPPLGLLPSLKELSIKGLDGIVSINADFFGSSSCS--FTSLKSLEFYHMKEWEEWECKGV 861
Query: 645 LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDR 704
FP L+ L I CPKLK LP+ L L L I+ C+ L S + +I L + C
Sbjct: 862 TGAFPRLQRLSIERCPKLKGHLPEQLCHLNSLKISGCEQLVPSALSAPDIHKLYLGDCGE 921
Query: 705 ILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDL-CCYNSL 763
+ ++ T+LK E ++E V + + E E+ N S + + CY+ L
Sbjct: 922 LQIDH-GTTLK----------ELTIEGHNVEAALFE--EIGRNYSCSNNNIPMHSCYDFL 968
Query: 764 GELSITRWCSSSLSFSLHLFT-----------------------NLYSLWFVDCPNLDSF 800
L I C S +F L +FT +L +L +CP L+S
Sbjct: 969 VSLRIKGGCDSLTTFPLDMFTILRELCIWKCPNLRRISQGQAHNHLQTLDIKECPQLESL 1028
Query: 801 PEG--GLPCNLLSLTITNCPK--------LIASRQEWGLKSLKYFFVC---------DDF 841
PEG L +L SL I +CPK L ++ +E GL Y +
Sbjct: 1029 PEGMHVLLPSLDSLCIDDCPKVEMFPEGGLPSNLKEMGLFGGSYKLISLLKSALGGNHSL 1088
Query: 842 ENV-------ESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLE 894
E + E P+E +LP +L L +N+C L+ ++ +G HL SL+ L + +CP L+
Sbjct: 1089 ERLVIGKVDFECLPEEGVLPHSLVSLQINSCGDLKRLDYKGICHLSSLKELSLEDCPRLQ 1148
Query: 895 RLPEEALPNSLYSLWI-KDCPLIKVKYQKEGGEQRDTICH 933
LPEE LP S+ +LWI DC L+K + ++ GE I H
Sbjct: 1149 CLPEEGLPKSISTLWIWGDCQLLKQRCREPEGEDWPKIAH 1188
>UniRef100_Q6SQJ0 NBS-LRR type disease resistance protein Hom-F [Glycine max]
Length = 1124
Score = 563 bits (1452), Expect = e-159
Identities = 374/956 (39%), Positives = 534/956 (55%), Gaps = 86/956 (8%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGKK 59
+ND +I+ F++KAWV VS+ FDV +T+ IL++ S D + +Q +L+ L G K
Sbjct: 226 FNDPRIENKFDIKAWVCVSDEFDVFNVTRTILEAVTKSTDDSRNRETVQGRLREKLTGNK 285
Query: 60 YLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKS 119
+ LVLDD+WN + + W+ L P N+G+ GSKIVVTTR+K+VA +++ S + L+ L
Sbjct: 286 FFLVLDDVWNRNQKEWKDLQTPLNYGASGSKIVVTTRDKKVA-SIVGSNKTHCLELLQDD 344
Query: 120 NCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINIL 179
+CW LF HAF+ S P+ + +G KIVEKC GLPLA+ ++G LL + S EW IL
Sbjct: 345 HCWRLFTKHAFRDDSHQPNPDFKEIGTKIVEKCKGLPLALTTIGSLLHQKSSISEWEGIL 404
Query: 180 ETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLL 239
++++W S+ D ++ L LSYH+LPS+LKRCF+YC++FPK ++F ++ LI LWMAE L
Sbjct: 405 KSEIWEFSEEDSSIVPALALSYHHLPSHLKRCFAYCALFPKDYRFDEEGLIQLWMAENFL 464
Query: 240 KCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH--YVMHDLVNDLTKSVSGEFS 297
+C +RS E+ G + F DL+S SFFQQS T E +VMHDL+NDL K V G+
Sbjct: 465 QCHQQSRSPEKVGEQYFNDLLSRSFFQQS-----STVERTPFVMHDLLNDLAKYVCGDIC 519
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSV---DKLLEL-TCEGLHSLILEGTRAMLISNNV- 352
++E+ + + TRH FS+ S+ V D L E L + + + N+
Sbjct: 520 FRLENDQATNIPKTTRH--FSVASDHVTCFDGFRTLYNAERLRTFMSLSEEMSFRNYNLW 577
Query: 353 -----QQDLFSRLNFLRMLSFRG-CGLLELVDEISNLKLLRYLDLSYTWIEILPDTICML 406
++LFS+ FLR+LS G L ++ + + NLK L LDLS+T I LP++IC L
Sbjct: 578 YCKMSTRELFSKFKFLRVLSLSGYSNLTKVPNSVGNLKYLSSLDLSHTEIVKLPESICSL 637
Query: 407 HNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL-SY 465
+NLQ L L GC L ELPSN KL +L L+L ++ +P H GKL LQ L S
Sbjct: 638 YNLQILKLNGCEHLKELPSNLHKLTDLHRLELIDTE----VRKVPAHLGKLKYLQVLMSS 693
Query: 466 FIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGR 525
F V + +++L +LN LHG++ I L NV + +D+ V+LK+ +L EL +++D
Sbjct: 694 FNVGKSREFSIQQLGELN-LHGSLSIRQLQNVENPSDALAVDLKNKTHLVELELEWDSDW 752
Query: 526 EEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLC 585
D+S E + V+E LQP+++L++LT+S Y G FP W+ L +VSL L+ C
Sbjct: 753 NP-DDSTKERD--VIENLQPSKHLEKLTMSNYGGKQFPRWLFNNSLLRVVSLTLKNCKGF 809
Query: 586 SLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC- 644
LPPLG LP LK LSI DGI I +F SSS + F SLE L+F M WEEW C
Sbjct: 810 LCLPPLGRLPSLKELSIEGLDGIVSINADFLGSSSCS--FTSLESLEFSDMKEWEEWECK 867
Query: 645 --LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRC 702
FP L+ L I CPKLK LP+ L L L I+ L I LDI
Sbjct: 868 GVTGAFPRLRRLSIERCPKLKGHLPEQLCHLNSLKISGWDSL--------TTIPLDI--- 916
Query: 703 DRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNS 762
P LK+L I E ++++I + + L + +CP L+
Sbjct: 917 -------FPI-LKELQIWECP----NLQRISQGQALNHLETLSMR---ECPQLE------ 955
Query: 763 LGELSITRWCSSSLSFSLH-LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTI-TNCPKL 820
SL +H L +L SLW DCP ++ FPEGGLP NL S+ + KL
Sbjct: 956 ------------SLPEGMHVLLPSLDSLWIKDCPKVEMFPEGGLPSNLKSMGLYGGSYKL 1003
Query: 821 IASRQE--WGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLH 878
I+ + G SL+ + +VE P E +LP +L L + C L+ ++ G H
Sbjct: 1004 ISLLKSALGGNHSLERLVI--GGVDVECLPDEGVLPHSLVNLWIRECGDLKRLDYRGLCH 1061
Query: 879 LKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHI 934
L SL+ L + +CP LE LPEE LP S+ +L I +CPL+K + ++ GE I HI
Sbjct: 1062 LSSLKTLTLWDCPRLECLPEEGLPKSISTLGILNCPLLKQRCREPEGEDWPKIAHI 1117
>UniRef100_Q8GZF0 Resistance protein KR4 [Glycine max]
Length = 1211
Score = 563 bits (1450), Expect = e-158
Identities = 378/1013 (37%), Positives = 543/1013 (53%), Gaps = 103/1013 (10%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGKK 59
YND +I F++K W+ VSE FDV +++AIL + SAD G +L ++Q +L+ L KK
Sbjct: 219 YNDPRIVSMFDVKGWICVSEEFDVFNVSRAILDTITDSADDGRELEIVQRRLKERLADKK 278
Query: 60 YLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKS 119
+LLVLDD+WN +WE + +G+ GSKI+VTTR +EVA + ++ L+QL +
Sbjct: 279 FLLVLDDVWNESGPKWEAVQNALVYGAQGSKILVTTRSEEVASTM--GSDKHKLEQLQEG 336
Query: 120 NCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINIL 179
CW LF HAF+ ++ P + ++IVEKC GLPLA+KS+G LL EW ++L
Sbjct: 337 YCWELFAKHAFRDDNLPRDPVCTDISKEIVEKCRGLPLALKSMGSLLHNK-PAWEWESVL 395
Query: 180 ETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLL 239
++++W L D + L LSYH+LP +LK CF+YC++FPK + F ++ LI LWMAE L
Sbjct: 396 KSEIWELKNSD--IVPALALSYHHLPPHLKTCFAYCALFPKDYVFDRECLIQLWMAENFL 453
Query: 240 KCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQ 299
C + S EE G + F DL+S SFFQQ+ Y+ E +VMHDL+NDL K V G+ +
Sbjct: 454 NCHQCSTSPEEVGQQYFNDLLSRSFFQQASQ--YE--EGFVMHDLLNDLAKYVCGDIYFR 509
Query: 300 IEDARVERSVERTRHIWFSLQSNSVDKLLELTCEGLHSLILEGTRAMLISNNVQ------ 353
+ + + + + TRH S+ + +C+ T + N+
Sbjct: 510 LGVDQAKCTQKTTRHFSVSMITKPYFDEFGTSCDTKKLRTFMPTSWTMNENHSSWSCKMS 569
Query: 354 -QDLFSRLNFLRMLSFRGC-GLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHNLQT 411
+LFS+L FLR+LS C + EL D + N K LR LDLS T I+ LP++ C L+NLQ
Sbjct: 570 IHELFSKLKFLRVLSLSHCLDIKELPDSVCNFKHLRSLDLSETGIKKLPESTCSLYNLQI 629
Query: 412 LLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQ-SLSYFIVEE 470
L L C L ELPSN +L NL L+ + I MP H GKL NLQ S+S F V +
Sbjct: 630 LKLNHCRSLKELPSNLHELTNLHRLEFVNTE----IIKMPPHLGKLKNLQVSMSSFNVGK 685
Query: 471 QNVSDLKELAKLN-HLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMD 529
++ +++ +LN LH + L N+ + +D+ +LK+ L EL +++ R D
Sbjct: 686 RSEFTIQKFGELNLVLHERLSFRELQNIENPSDALAADLKNKTRLVELKFEWNSHRNP-D 744
Query: 530 ESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLP 589
+S E +V V+E LQP+++L++L+I Y G FPNW+ L N+ SL L C C LP
Sbjct: 745 DSAKERDVIVIENLQPSKHLEKLSIRNYGGKQFPNWLSDNSLSNVESLVLDNCQSCQRLP 804
Query: 590 PLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC---LE 646
LG LPFL+ L IS DGI IG +F+ +S+ + F SLE LKF M WE+W C
Sbjct: 805 SLGLLPFLENLEISSLDGIVSIGADFHGNSTSS--FPSLERLKFSSMKAWEKWECEAVTG 862
Query: 647 GFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDRIL 706
FP LK L I +CPKLK LP+ L L+KL I++CK LEAS P + L++++ D
Sbjct: 863 AFPCLKYLSISKCPKLKGDLPEQLLPLKKLKISECKQLEASAPRALE-LKLELEQQDFGK 921
Query: 707 VNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLE----------------------- 743
+ +LK L + Y+ + + V S LE L+
Sbjct: 922 LQLDWATLKTLSM--RAYSNYKEALLLVKSDTLEELKIYCCRKDGMDCDCEMRDDGCDSQ 979
Query: 744 -------------LDLNG--SLKCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYS 788
L+LNG +L+ T D +N L L+I R SL T+L
Sbjct: 980 KTFPLDFFPALRTLELNGLRNLQMITQDQ-THNHLEFLTIRRCPQLE---SLPGSTSLKE 1035
Query: 789 LWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGL---KSLKYFFVCDDFENVE 845
L DCP ++SFPEGGLP NL + + C + + + L SLK + ++ E
Sbjct: 1036 LAICDCPRVESFPEGGLPSNLKEMHLYKCSSGLMASLKGALGDNPSLKTLRIIK--QDAE 1093
Query: 846 SFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHL-----------------------KSL 882
SFP E LLP +L+ L + + L+ ++ +G HL KS+
Sbjct: 1094 SFPDEGLLPLSLACLVIRDFPNLKKLDYKGLCHLSSLKKLILDYCPNLQQLPEEGLPKSI 1153
Query: 883 EFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHIP 935
FL I CP+L++LPEE LP S+ L IK CP +K + Q GGE I HIP
Sbjct: 1154 SFLSIEGCPNLQQLPEEGLPKSISFLSIKGCPKLKQRCQNPGGEDWPKIAHIP 1206
>UniRef100_Q6SQI9 NBS-LRR type disease resistance protein Hom-B [Glycine max]
Length = 1124
Score = 561 bits (1446), Expect = e-158
Identities = 376/962 (39%), Positives = 538/962 (55%), Gaps = 98/962 (10%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGKK 59
+ND +I+ F++KAWV VS+ FDV +T+ IL++ S D + +Q +L+ L G K
Sbjct: 226 FNDPRIENKFDIKAWVCVSDEFDVFNVTRTILEAVTKSTDDSRNRETVQGRLREKLTGNK 285
Query: 60 YLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKS 119
+ LVLDD+WN + + W+ L P N+G+ GSKIVVTTR+K+VA +++ S + L+ L
Sbjct: 286 FFLVLDDVWNRNQKEWKDLQTPLNYGASGSKIVVTTRDKKVA-SIVGSNKTHCLELLQDD 344
Query: 120 NCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINIL 179
+CW LF HAF+ S P+ + +G KIVEKC GLPLA+ ++G LL + S EW IL
Sbjct: 345 HCWRLFTKHAFRDDSHQPNPDFKEIGTKIVEKCKGLPLALTTIGSLLHQKSSISEWEGIL 404
Query: 180 ETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLL 239
++++W S+ D ++ L LSYH+LPS+LKRCF+YC++FPK ++F K+ LI LWMAE L
Sbjct: 405 KSEIWEFSEEDSSIVPALALSYHHLPSHLKRCFAYCALFPKDYRFDKEGLIQLWMAENFL 464
Query: 240 KCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH--YVMHDLVNDLTKSVSGEFS 297
+C +RS E+ G + F DL+S S FQQS T E +VMHDL+NDL K V G+
Sbjct: 465 QCHQQSRSPEKVGEQYFNDLLSRSLFQQS-----STVERTPFVMHDLLNDLAKYVCGDIC 519
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSVDKLLELTC-EGLHSLI-LEGTRA-MLISNNVQ- 353
++E+ + + TRH FS+ S+ V TC +G +L E R M +S +
Sbjct: 520 FRLENDQATNIPKTTRH--FSVASDHV------TCFDGFRTLYNAERLRTFMSLSEEMSF 571
Query: 354 ------------QDLFSRLNFLRMLSFRG-CGLLELVDEISNLKLLRYLDLSYTWIEILP 400
++LFS+ FLR+LS G L ++ + + NLK L LDLS+T I LP
Sbjct: 572 RNYNPWYCKMSTRELFSKFKFLRVLSLSGYYNLTKVPNSVGNLKYLSSLDLSHTEIVKLP 631
Query: 401 DTICMLHNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNL 460
++IC L+NLQ L L GC L ELPSN KL +L L+L ++ +P H GKL L
Sbjct: 632 ESICSLYNLQILKLNGCEHLKELPSNLHKLTDLHRLELIDTE----VRKVPAHLGKLKYL 687
Query: 461 QSL-SYFIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHM 519
Q L S F V + +++L +LN LHG++ I L NV + +D+ V+LK+ +L EL +
Sbjct: 688 QVLMSSFNVGKSREFSIQQLGELN-LHGSLSIRQLQNVENPSDALAVDLKNKTHLVELEL 746
Query: 520 KFDGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNL 579
++D D+S E + V+E LQP+++L++LT+S Y G FP W+ L +VSL L
Sbjct: 747 EWDSDWNP-DDSTKERD--VIENLQPSKHLEKLTMSNYGGKQFPRWLFNNSLLRVVSLTL 803
Query: 580 QFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNW 639
+ C LPPLG LP LK LSI DGI I +F+ SSS + F SLE L+F M W
Sbjct: 804 KNCKGFLCLPPLGRLPSLKELSIEGLDGIVSINADFFGSSSCS--FTSLESLEFSDMKEW 861
Query: 640 EEWLC---LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIID 696
EEW C FP L+ L I CPKLK LP+ L L L I+ L I
Sbjct: 862 EEWECKGVTGAFPRLQRLSIMRCPKLKGHLPEQLCHLNYLKISGWDSL--------TTIP 913
Query: 697 LDIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLD 756
LDI P LK+L I E ++++I + + L + +CP L+
Sbjct: 914 LDI----------FPI-LKELQIWECP----NLQRISQGQALNHLETLSMR---ECPQLE 955
Query: 757 LCCYNSLGELSITRWCSSSLSFSLH-LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTI- 814
SL +H L +L SLW DCP ++ FPEGGLP NL S+ +
Sbjct: 956 ------------------SLPEGMHVLLPSLDSLWIDDCPKVEMFPEGGLPSNLKSMGLY 997
Query: 815 TNCPKLIASRQE--WGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMN 872
KLI+ + G SL+ + +VE P E +LP +L L + C L+ ++
Sbjct: 998 GGSYKLISLLKSALGGNHSLERLVI--GGVDVECLPDEGVLPHSLVNLWIRECGDLKRLD 1055
Query: 873 NEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTIC 932
+G HL SL+ L + +CP L+ LPEE LP S+ +L I +CPL+K + ++ GE I
Sbjct: 1056 YKGLCHLSSLKTLTLWDCPRLQCLPEEGLPKSISTLGILNCPLLKQRCREPEGEDWPKIA 1115
Query: 933 HI 934
HI
Sbjct: 1116 HI 1117
>UniRef100_Q6R271 Disease resistance protein [Glycine max]
Length = 1129
Score = 561 bits (1446), Expect = e-158
Identities = 376/962 (39%), Positives = 538/962 (55%), Gaps = 98/962 (10%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSAD-GEDLNLLQHQLQYMLMGKK 59
+ND +I+ F++KAWV VS+ FDV +T+ IL++ S D + +Q +L+ L G K
Sbjct: 226 FNDPRIENKFDIKAWVCVSDEFDVFNVTRTILEAVTKSTDDSRNRETVQGRLREKLTGNK 285
Query: 60 YLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKS 119
+ LVLDD+WN + + W+ L P N+G+ GSKIVVTTR+K+VA +++ S + L+ L
Sbjct: 286 FFLVLDDVWNRNQKEWKDLQTPLNYGASGSKIVVTTRDKKVA-SIVGSNKTHCLELLQDD 344
Query: 120 NCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINIL 179
+CW LF HAF+ S P+ + +G KIVEKC GLPLA+ ++G LL + S EW IL
Sbjct: 345 HCWRLFTKHAFRDDSHQPNPDFKEIGTKIVEKCKGLPLALTTIGSLLHQKSSISEWEGIL 404
Query: 180 ETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLL 239
++++W S+ D ++ L LSYH+LPS+LKRCF+YC++FPK ++F K+ LI LWMAE L
Sbjct: 405 KSEIWEFSEEDSSIVPALALSYHHLPSHLKRCFAYCALFPKDYRFDKEGLIQLWMAENFL 464
Query: 240 KCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH--YVMHDLVNDLTKSVSGEFS 297
+C +RS E+ G + F DL+S S FQQS T E +VMHDL+NDL K V G+
Sbjct: 465 QCHQQSRSPEKVGEQYFNDLLSRSLFQQS-----STVERTPFVMHDLLNDLAKYVCGDIC 519
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSVDKLLELTC-EGLHSLI-LEGTRA-MLISNNVQ- 353
++E+ + + TRH FS+ S+ V TC +G +L E R M +S +
Sbjct: 520 FRLENDQATNIPKTTRH--FSVASDHV------TCFDGFRTLYNAERLRTFMSLSEEMSF 571
Query: 354 ------------QDLFSRLNFLRMLSFRG-CGLLELVDEISNLKLLRYLDLSYTWIEILP 400
++LFS+ FLR+LS G L ++ + + NLK L LDLS+T I LP
Sbjct: 572 RNYNPWYCKMSTRELFSKFKFLRVLSLSGYYNLTKVPNSVGNLKYLSSLDLSHTEIVKLP 631
Query: 401 DTICMLHNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNL 460
++IC L+NLQ L L GC L ELPSN KL +L L+L ++ +P H GKL L
Sbjct: 632 ESICSLYNLQILKLNGCEHLKELPSNLHKLTDLHRLELIDTE----VRKVPAHLGKLKYL 687
Query: 461 QSL-SYFIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHM 519
Q L S F V + +++L +LN LHG++ I L NV + +D+ V+LK+ +L EL +
Sbjct: 688 QVLMSSFNVGKSREFSIQQLGELN-LHGSLSIRQLQNVENPSDALAVDLKNKTHLVELEL 746
Query: 520 KFDGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNL 579
++D D+S E + V+E LQP+++L++LT+S Y G FP W+ L +VSL L
Sbjct: 747 EWDSDWNP-DDSTKERD--VIENLQPSKHLEKLTMSNYGGKQFPRWLFNNSLLRVVSLTL 803
Query: 580 QFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNW 639
+ C LPPLG LP LK LSI DGI I +F+ SSS + F SLE L+F M W
Sbjct: 804 KNCKGFLCLPPLGRLPSLKELSIEGLDGIVSINADFFGSSSCS--FTSLESLEFSDMKEW 861
Query: 640 EEWLC---LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIID 696
EEW C FP L+ L I CPKLK LP+ L L L I+ L I
Sbjct: 862 EEWECKGVTGAFPRLQRLSIMRCPKLKGHLPEQLCHLNYLKISGWDSL--------TTIP 913
Query: 697 LDIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLD 756
LDI P LK+L I E ++++I + + L + +CP L+
Sbjct: 914 LDI----------FPI-LKELQIWECP----NLQRISQGQALNHLETLSMR---ECPQLE 955
Query: 757 LCCYNSLGELSITRWCSSSLSFSLH-LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTI- 814
SL +H L +L SLW DCP ++ FPEGGLP NL S+ +
Sbjct: 956 ------------------SLPEGMHVLLPSLDSLWIDDCPKVEMFPEGGLPSNLKSMGLY 997
Query: 815 TNCPKLIASRQE--WGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMN 872
KLI+ + G SL+ + +VE P E +LP +L L + C L+ ++
Sbjct: 998 GGSYKLISLLKSALGGNHSLERLVI--GGVDVECLPDEGVLPHSLVNLWIRECGDLKRLD 1055
Query: 873 NEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTIC 932
+G HL SL+ L + +CP L+ LPEE LP S+ +L I +CPL+K + ++ GE I
Sbjct: 1056 YKGLCHLSSLKTLTLWDCPRLQCLPEEGLPKSISTLGILNCPLLKQRCREPEGEDWPKIA 1115
Query: 933 HI 934
HI
Sbjct: 1116 HI 1117
>UniRef100_Q93VS9 NBS-LRR resistance-like protein B8 [Phaseolus vulgaris]
Length = 1133
Score = 543 bits (1399), Expect = e-153
Identities = 357/954 (37%), Positives = 513/954 (53%), Gaps = 70/954 (7%)
Query: 1 YNDNKIKE-HFELKAWVYVSESFDVVGLTKAILKSFNSSA-DGEDLNLLQHQLQYMLMGK 58
Y+D KI++ F++KAWV VS+ F V+ +T+ IL++ D +L ++ +L+ L+GK
Sbjct: 225 YSDPKIEDAKFDIKAWVCVSDHFHVLTVTRTILEAITDKTNDSGNLEMVHKKLKEKLLGK 284
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
++LLVLDD+WN WE + P ++G+ GS+I+VTTR ++VA ++ L L+QL +
Sbjct: 285 RFLLVLDDVWNERPAEWEAVRTPLSYGAPGSRILVTTRSEKVASSMRSEVHL--LKQLGE 342
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
CW +F HA + + L VGR+IVEKC GLPLA+K++G LL S +W NI
Sbjct: 343 DECWKVFENHALKDGDLELNDELMKVGRRIVEKCKGLPLALKTIGCLLSTKSSISDWKNI 402
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
LE+D+W+L K + L LSY +LPS+LKRCF+YC++FPK ++F K+ELI LWMA+
Sbjct: 403 LESDIWKLPKEHSEIIPALFLSYRHLPSHLKRCFAYCALFPKDYEFVKEELIFLWMAQNF 462
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
L R EE G E F DL+S FF QS H+VMHDL+NDL K V +F
Sbjct: 463 LLSPQHIRDPEEIGEEYFNDLLSRCFFNQS-----SIVGHFVMHDLLNDLAKYVCADFCF 517
Query: 299 QIEDARVERSVERTRHIWFS-LQSNSVDKLLELT-CEGLHSL--ILEGTRAMLISNNVQQ 354
+++ + + T H F L S D LT + L S I E A
Sbjct: 518 RLKFDNEKCMPKTTCHFSFEFLDVESFDGFESLTNAKRLRSFLPISETWGASWHFKISIH 577
Query: 355 DLFSRLNFLRMLSFRGC-GLLELVDEISNLKLLRYLDLSYTWIEILPDTICMLHNLQTLL 413
DLFS++ F+R+LSF GC L E+ D + +LK L+ LDLS T I+ LPD+IC+L+NL L
Sbjct: 578 DLFSKIKFIRVLSFHGCLDLREVPDSVGDLKHLQSLDLSSTEIQKLPDSICLLYNLLILK 637
Query: 414 LEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVEEQNV 473
L C +L E P N KL LR L+ + ++ MP H G+L NLQ LS F+V++ +
Sbjct: 638 LSSCSKLKEFPLNLHKLTKLRCLEFEGTD----VRKMPMHFGELKNLQVLSMFLVDKNSE 693
Query: 474 SDLKELAKLN--HLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDES 531
K+L L +LHG + I + N+ + D+ NLKD K L +L +K+ D
Sbjct: 694 LSTKQLGGLGGLNLHGRLSINDVQNIGNPLDALKANLKD-KRLVKLELKWKWNHVPDDPK 752
Query: 532 MAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPL 591
+ VL+ LQP+ +L++L I Y G FP+W+ L NLV LNL+ C C LP L
Sbjct: 753 KEKE---VLQNLQPSNHLEKLLIRNYSGTEFPSWVFDNSLSNLVFLNLEDCKYCLCLPSL 809
Query: 592 GTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC-LEGFPL 650
G L LK+L IS DGI IG EFY S+S F SLE L+F M WEEW C FP
Sbjct: 810 GLLSSLKILHISGLDGIVSIGAEFYGSNSS---FASLERLEFHNMKEWEEWECKTTSFPR 866
Query: 651 LKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDRILVNEL 710
L+ LY+ +CPKLK + K+ ++D + + +D D I
Sbjct: 867 LEVLYVDKCPKLKGT---------KVVVSDELRISGN--------SMDTSHTDGIFRLHF 909
Query: 711 PTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGELSITR 770
L+ L LE+ + Q + ++ ++ + D CP + +
Sbjct: 910 FPKLRSLQ-LEDCQNLRRISQEYAHNHLMNLYIHD------CPQFKSFLFPKPSLTKLKS 962
Query: 771 WCSSSLSFSLH------LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASR 824
+ S L L LF +L L V CP ++ FP+GGLP N+ +++++ +++ R
Sbjct: 963 FLFSELKSFLFPKPMQILFPSLTELHIVKCPEVELFPDGGLPLNIKHISLSSLKLIVSLR 1022
Query: 825 QEW----GLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLK 880
L+SL ++ VE FP E LLP +L+ L + C L+ M+ +G HL
Sbjct: 1023 DNLDPNTSLQSLNIHYL-----EVECFPDEVLLPRSLTSLGIRWCPNLKKMHYKGLCHLS 1077
Query: 881 SLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHI 934
SL L CPSL+ LP E LP S+ SL I CPL+K + + GE I HI
Sbjct: 1078 SLTLL---ECPSLQCLPTEGLPKSISSLTICGCPLLKERCRNPDGEDWRKIAHI 1128
>UniRef100_Q93VN8 NBS-LRR resistance-like protein B11 [Phaseolus vulgaris]
Length = 1105
Score = 538 bits (1386), Expect = e-151
Identities = 359/953 (37%), Positives = 515/953 (53%), Gaps = 90/953 (9%)
Query: 1 YNDNKIKE-HFELKAWVYVSESFDVVGLTKAILKSF-NSSADGEDLNLLQHQLQYMLMGK 58
Y+D KI++ F++KAWV VS+ F V+ +T+ IL++ N + D +L ++ +L+ L+GK
Sbjct: 218 YSDPKIEDAKFDIKAWVCVSDHFHVLTVTRTILEAITNQNDDSGNLEMVHKKLKEKLLGK 277
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
++LLVLDD+WN WE + P ++G+ GS+I+ TTR ++VA ++ L L+QL +
Sbjct: 278 RFLLVLDDVWNERPAEWEAVRTPLSYGAPGSRILFTTRSEKVASSMRSEVHL--LKQLGE 335
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
CW +F HA + + L VGR+IVEKC GLPLA+K++G LL S +W NI
Sbjct: 336 DECWKVFENHALKDGDLELNDELMKVGRRIVEKCKGLPLALKTIGCLLSTKSSISDWKNI 395
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
LE+D+W L K + L LSY +LPS+LKRCF+YC++FPK +KF K+ELI LWMA+
Sbjct: 396 LESDIWELPKEHSEIIPALFLSYRHLPSHLKRCFAYCALFPKDYKFVKEELIFLWMAQNF 455
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
L R EE G E F DL+S FF QS +VMHDL+NDL K V +F
Sbjct: 456 LLSPQQIRHPEEVGEEYFNDLLSRCFFNQS-----SFVGRFVMHDLLNDLAKYVCADFCF 510
Query: 299 QIEDARVERSVERTRHIWFSLQSNSVDKLLELTCEGLHSLI-LEGTRAMLISNNVQQ--- 354
+++ + + + TRH FS + V+ +G SL + R+ L + + +
Sbjct: 511 RLKYDKCQCIPKTTRH--FSFEFRDVESF-----DGFESLTDAKRLRSFLPISKLWEPKW 563
Query: 355 -------DLFSRLNFLRMLSFRGC-GLLELVDEISNLKLLRYLDLSYTWIEILPDTICML 406
DLFS++ F+R+LSF GC L E+ D + +LK L+ LDLS+T I LP++IC+L
Sbjct: 564 HFKISIHDLFSKIKFIRVLSFNGCLDLREVPDSVGDLKHLQSLDLSWTMIRKLPNSICLL 623
Query: 407 HNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYF 466
+NL L L C L E P N KL LR L+ ++ MP H G+L NLQ LS F
Sbjct: 624 YNLLILKLNSCSVLMEFPLNLHKLTKLRCLEFKG----TMVRKMPMHFGELKNLQVLSKF 679
Query: 467 IVEEQNVSDLKELAKLN--HLHGAIDIEGLGNVSDLADSATVNLKDTKYLE-ELHMKFDG 523
V++ + KEL L +LHG + I + N+ + D+ NLKD + +E EL K D
Sbjct: 680 FVDKNSELSTKELGGLGGLNLHGRLSINDVQNIGNPLDALKANLKDKRLVELELQWKSDH 739
Query: 524 GREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCG 583
++ + VL+ LQP+ +L++L+I Y G FP+W + NLV L L C
Sbjct: 740 ITDD-----PKKEKEVLQNLQPSIHLEKLSIISYNGREFPSW--EFDNSNLVILKLANCK 792
Query: 584 LCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWL 643
C LPPLG L LK L I DGI +G+EFY S+S F SLE L F M WEEW
Sbjct: 793 YCLCLPPLGLLSSLKTLEIIGLDGIVSVGDEFYGSNS---SFASLERLYFLNMKEWEEWE 849
Query: 644 C-LEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRC 702
C FP L+ELY+ CPKLK + + +L I+ M + G + K C
Sbjct: 850 CETTSFPRLEELYVGGCPKLKGT---KVVVSDELRISGNSMDTSHTDGGSFRLHFFPKLC 906
Query: 703 DRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNS 762
L++ +LK+ + Q VN+ ++++ CP L +
Sbjct: 907 TLKLIH--CQNLKR------------ISQESVNNHLIQL------SIFSCPQLKSFLFPK 946
Query: 763 LGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIA 822
++ LF +L L C ++ FP+GGLP N+ +++ +C KLIA
Sbjct: 947 PMQI---------------LFPSLTKLEISKCAEVELFPDGGLPLNIKEMSL-SCLKLIA 990
Query: 823 S-RQEWGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKS 881
S R + DD E VE FP E LLP +L+ L + C L+ M+ +G HL S
Sbjct: 991 SLRDNLDPNTSLQSLTIDDLE-VECFPDEVLLPRSLTSLYIEYCPNLKKMHYKGLCHLSS 1049
Query: 882 LEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHI 934
LE ++NCPSLE LP E LP S+ SL I +CPL+K + Q GE + I HI
Sbjct: 1050 LE---LLNCPSLECLPAEGLPKSISSLTIFNCPLLKERCQSPDGEDWEKIAHI 1099
>UniRef100_Q93W58 NBS-LRR resistance-like protein J71 [Phaseolus vulgaris]
Length = 1066
Score = 519 bits (1336), Expect = e-145
Identities = 348/947 (36%), Positives = 501/947 (52%), Gaps = 124/947 (13%)
Query: 1 YNDNKIK-EHFELKAWVYVSESFDVVGLTKAILKSF-NSSADGEDLNLLQHQLQYMLMGK 58
YND KI F++KAWV VS+ F V+ +T+ IL++ N D +L ++ +L+ L G+
Sbjct: 223 YNDRKIDGAKFDIKAWVCVSDHFHVLTVTRTILEAITNQKDDSGNLEMVHKKLKEKLSGR 282
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
K+ LVLDD+WN E WE++ P ++G+ GSKI+VTTRE++VA N+ S+++ L+QL +
Sbjct: 283 KFFLVLDDVWNEKREEWEVVRTPLSYGAPGSKILVTTREEKVASNM--SSKVHRLKQLRE 340
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
CW++F HA + L+ +GR+IV++C GLPLA+K++G LLR S +W NI
Sbjct: 341 EECWNVFENHALKDGDYELNDELKEIGRRIVDRCKGLPLALKTIGCLLRTKSSISDWKNI 400
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
LE+++W L K ++ + L +SY LPS+LK+CF+YC++FPK ++F+K ELI++WMA+
Sbjct: 401 LESEIWELPKENNEIIPALFMSYRYLPSHLKKCFAYCALFPKDYEFEKKELILMWMAQNF 460
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
L+C R EE G E F DL+S SFFQQS ++MHDL+NDL K V +F
Sbjct: 461 LQCPQQVRHREEVGEEYFNDLLSRSFFQQS-----GVRRRFIMHDLLNDLAKYVCADFCF 515
Query: 299 QIEDARVERSVERTRHIWFSLQS-NSVDKLLELTCEGLHSLILEGTRAMLISNNVQ---Q 354
+++ + + + TRH F S D L+ L+ ++AM + N +
Sbjct: 516 RLKFDKGQCIPKTTRHFSFEFHDIKSFDGFGSLSDAKRLRSFLQFSQAMTLQWNFKISIH 575
Query: 355 DLFSRLNFLRMLSFRGCGLL-ELVDEISNLKLLRYLDLSY-TWIEILPDTICMLHNLQTL 412
DLFS++ F+RMLSF GC L E+ D + +LK L LDLS + I+ LPD+IC+L+NL L
Sbjct: 576 DLFSKIKFIRMLSFCGCSFLKEVPDSVGDLKHLHSLDLSACSAIKKLPDSICLLYNLLIL 635
Query: 413 LLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVEEQN 472
L C L ELP N KL LR L+ + MP H G+L NLQ L+ F V+ +
Sbjct: 636 KLNKCVNLKELPINLHKLTKLRCLEFEGTR----VSKMPMHFGELKNLQVLNPFFVDRNS 691
Query: 473 VSDLKELAKLNHLH--GAIDIEGLGNVSDLADSATVNLKDTKYLE-ELHMKFDGGREEMD 529
K+LA L L+ + I L N+ + D+ N+KD +E EL K+D D
Sbjct: 692 ELIPKQLAGLGGLNIQKRLSINDLQNILNPLDALKANVKDKDLVELELKWKWD---HIPD 748
Query: 530 ESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLP 589
+ E V L+ LQP+++L+ L+I Y G FP+W+ L NLV L L C C P
Sbjct: 749 DPRKEKEV--LQNLQPSKHLEGLSIRNYSGTEFPSWVFDNSLSNLVFLELNNCKYCLCFP 806
Query: 590 PLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEGFP 649
PLG L LK L I DGI IG EFY S+S F SLE L+F M WEEW C
Sbjct: 807 PLGLLSSLKTLGIVGLDGIVSIGAEFYGSNSS---FASLERLEFHDMKEWEEWECKT--- 860
Query: 650 LLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGDNIIDLDIKRCDRILVNE 709
P LQ+L + +C L+
Sbjct: 861 ------------------TSFPRLQELSVIECPKLKG----------------------- 879
Query: 710 LPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGELSIT 769
T LKK+F+ E EL ++G+ ++
Sbjct: 880 --THLKKVFVSE---------------------ELTISGN---------------SMNTD 901
Query: 770 RWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEW-- 827
C S F L F L+SL + C N+ P N+ ++++ C KLIAS ++
Sbjct: 902 GGCDSLTIFRLDFFPKLFSLELITCQNIRRIS----PLNIKEMSLS-CLKLIASLRDNLD 956
Query: 828 GLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYI 887
SL+ F+ D VE FP E LLP +L+ L+++ C L+ M+ +G HL SL +
Sbjct: 957 PNTSLESLFIFD--LEVECFPDEVLLPRSLTSLDISFCRNLKKMHYKGLCHLSSLT---L 1011
Query: 888 INCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHI 934
+CPSLE LP E LP S+ SL I+DCPL+K + + GE I HI
Sbjct: 1012 YDCPSLECLPAEGLPKSISSLTIRDCPLLKERCRNPDGEDWGKIAHI 1058
>UniRef100_Q60D45 Putative disease resistance protein I2 [Solanum demissum]
Length = 1190
Score = 516 bits (1329), Expect = e-144
Identities = 338/1007 (33%), Positives = 531/1007 (52%), Gaps = 115/1007 (11%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKS---FNSSADGEDLNLLQHQLQYMLMG 57
YND ++K HF LKAW VSE +D + +TK +L+ F+S +LN LQ +L+ L
Sbjct: 222 YNDERVKNHFGLKAWYCVSEGYDALRITKGLLQEIGKFDSKDVHNNLNQLQVKLKESLKE 281
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
KK+L+VLDD+WN + W+ L F G GSKI+VTTR++ VA ++ E + L
Sbjct: 282 KKFLIVLDDVWNDNYNEWDDLRNTFVQGDIGSKIIVTTRKESVA--LMMGNEQISMDNLS 339
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
WSLF HAF+ +P LE VG +I KC GLPLA+K+L +LR EW
Sbjct: 340 TEASWSLFKRHAFENMDPMGHPELEEVGNQIAAKCKGLPLALKTLAGMLRSKSEVEEWKR 399
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
IL +++W L D + L LSY++LP++LKRCFSYC+IFPK + F+K+++I LW+A G
Sbjct: 400 ILRSEIWELPHND--IVPALMLSYNDLPAHLKRCFSYCAIFPKDYSFRKEQVIHLWIANG 457
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFD-EIYDTYEHYVMHDLVNDLTKSVSGEF 296
L++ + E+ GN+ F +L S S F++ + + + E ++MHDL+NDL + S +
Sbjct: 458 LVQ--KEDEIIEDSGNQYFLELRSRSLFEKVPNPSVGNIEELFLMHDLINDLAQIASSKL 515
Query: 297 SIQIEDARVERSVERTRHIWFSL-QSNSVDKLLEL-TCEGLHSL--ILEGTRAMLISNNV 352
I++E+++ +E++RH+ +S+ + +KL L E L +L I +S V
Sbjct: 516 CIRLEESQGSHMLEKSRHLSYSMGEGGEFEKLTTLYKLEQLRTLLPIYIDVNYYSLSKRV 575
Query: 353 QQDLFSRLNFLRMLSFRGCGLLELVDEIS-NLKLLRYLDLSYTWIEILPDTICMLHNLQT 411
++ RL LR+LS + EL +++ LKLLR+LD+S T I+ LPD+IC+L+NL+T
Sbjct: 576 LYNILPRLRSLRVLSLSYYNIKELPNDLFIELKLLRFLDISRTKIKRLPDSICVLYNLET 635
Query: 412 LLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVE 469
LLL C +L ELP KL+NLRHL + + + + MP H KL +LQ L + F++
Sbjct: 636 LLLSSCADLEELPLQMEKLINLRHLDISNTS----LLKMPLHLSKLKSLQVLVGAKFLLS 691
Query: 470 EQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMD 529
+ DL E +L+G++ + L NV D ++ +++ ++++L +++ E
Sbjct: 692 GWRMEDLGEA---QNLYGSVSVVELENVVDRREAVKAKMREKNHVDKLSLEWS---ESSS 745
Query: 530 ESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLP 589
+++ +L+ L+P++N+K + I+ Y+G FPNW+ LV L++ C C LP
Sbjct: 746 ADNSQTERDILDELRPHKNIKEVEITGYRGTKFPNWLADPLFLKLVQLSIDNCKDCYTLP 805
Query: 590 PLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEG-- 647
LG LP LK LSIS GI + EEFY S S F LE L FE M W++W L
Sbjct: 806 ALGQLPCLKFLSISGMHGITEVTEEFYGSFSSKKPFNCLEKLAFEDMPEWKQWHVLGSGE 865
Query: 648 FPLLKELYIRECPKLKMSLPQHLPSLQKLFINDC----------KMLEASIPNGDNIIDL 697
FP+L++L+I+ CP+L + P L SL+ ++ C ++ + + I++L
Sbjct: 866 FPILEKLFIKNCPELSLETPIQLSSLKSFEVSGCPKVGVVFDDAQLFRSQLEGMKQIVEL 925
Query: 698 DIKRCDRIL---VNELPTSLKKLFI-------LENRYTEFS--VEQIFV-NSTILEVLEL 744
I C+ + + LPT+LK++ I LE E S +E++ V S ++V+
Sbjct: 926 YISYCNSVTFLPFSILPTTLKRIEISRCRKLKLEAPVGEMSMFLEELRVEGSDCIDVISP 985
Query: 745 DL----------------NGSLKCPTLDLCCYNSLGELSITRWCSSSLSFSL-------- 780
+L + T LC ++ ++ C +L SL
Sbjct: 986 ELLPRARNLRVVSCHNLTRVLIPTATAFLCIWDCENVEKLSVACGGTLMTSLTIGCCSKL 1045
Query: 781 --------HLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKSL 832
L +L L CP ++SFP+GGLP NL L I+ C KL+ R+EW L+ L
Sbjct: 1046 KCLPERMQELLPSLKELDLRKCPEIESFPQGGLPFNLQILEISECKKLVNGRKEWRLQRL 1105
Query: 833 KYFFV--CDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINC 890
+ C + +++ ES LP +LS L II C
Sbjct: 1106 SQLAIYGCPNLQSL----SESALPSSLSKLT-------------------------IIGC 1136
Query: 891 PSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHIPCV 937
P+L+ LP + +P+SL L I +CPL+ + + GE I P +
Sbjct: 1137 PNLQSLPVKGMPSSLSELHISECPLLTALLEFDKGEYWPNIAQFPTI 1183
>UniRef100_Q9XET3 Disease resistance protein I2 [Lycopersicon esculentum]
Length = 1266
Score = 515 bits (1327), Expect = e-144
Identities = 350/1005 (34%), Positives = 527/1005 (51%), Gaps = 106/1005 (10%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKS---FNSSADGEDLNLLQHQLQYMLMG 57
YND ++K HF+LKAW VSE FD + +TK +L+ F+S +LN LQ +L+ L G
Sbjct: 215 YNDERVKNHFDLKAWYCVSEGFDALRITKELLQEIGKFDSKDVHNNLNQLQVKLKESLKG 274
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
KK+L+VLDD+WN + W L F G GSKI+VTTR+ VA ++ E + L
Sbjct: 275 KKFLIVLDDVWNENYNEWNDLRNIFAQGDIGSKIIVTTRKDSVA--LMMGNEQIRMGNLS 332
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
WSLF HAF+ +P LE VGR+I KC GLPLA+K+L +LR EW
Sbjct: 333 TEASWSLFQRHAFENMDPMGHPELEEVGRQIAAKCKGLPLALKTLAGMLRSKSEVEEWKR 392
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
IL +++W L D + L LSY++LP++LKRCFS+C+IFPK + F+K+++I LW+A G
Sbjct: 393 ILRSEIWELPHND--ILPALMLSYNDLPAHLKRCFSFCAIFPKDYPFRKEQVIHLWIANG 450
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH-YVMHDLVNDLTKSVSGEF 296
L+ + ++ GN+ F +L S S F++ + E ++MHDLVNDL + S +
Sbjct: 451 LVPV--KDEINQDLGNQYFLELRSRSLFEKVPNPSKRNIEELFLMHDLVNDLAQLASSKL 508
Query: 297 SIQIEDARVERSVERTRHIWFSLQSNSVDKLLE--LTCEGLHSLILEGTRAML--ISNNV 352
I++E+++ +E+ RH+ +S+ N K L E L +L+ L +S V
Sbjct: 509 CIRLEESQGSHMLEQCRHLSYSIGFNGEFKKLTPLYKLEQLRTLLPIRIEFRLHNLSKRV 568
Query: 353 QQDLFSRLNFLRMLSFRGCGLLELVDEI-SNLKLLRYLDLSYTWIEILPDTICMLHNLQT 411
++ L LR LSF + EL +++ + LKLLR+LD+S TWI LPD+IC L+NL+T
Sbjct: 569 LHNILPTLRSLRALSFSQYKIKELPNDLFTKLKLLRFLDISRTWITKLPDSICGLYNLET 628
Query: 412 LLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVE 469
LLL C +L ELP KL+NLRHL + S+ R MP H +L +LQ L F V+
Sbjct: 629 LLLSSCADLEELPLQMEKLINLRHLDV-SNTRR---LKMPLHLSRLKSLQVLVGPKFFVD 684
Query: 470 EQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMD 529
+ DL E +LHG++ + L NV D ++ +++ ++E+L +++ D
Sbjct: 685 GWRMEDLGEA---QNLHGSLSVVKLENVVDRREAVKAKMREKNHVEQLSLEWSES-SIAD 740
Query: 530 ESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLP 589
S ES++ L+ L P++N+K++ IS Y+G +FPNW+ LV+L+L+ C C LP
Sbjct: 741 NSQTESDI--LDELCPHKNIKKVEISGYRGTNFPNWVADPLFLKLVNLSLRNCKDCYSLP 798
Query: 590 PLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEW--LCLEG 647
LG LP LK LS+ GI+++ EEFY S F SLE L+FE M W++W L +
Sbjct: 799 ALGQLPCLKFLSVKGMHGIRVVTEEFYGRLSSKKPFNSLEKLEFEDMTEWKQWHALGIGE 858
Query: 648 FPLLKELYIRECPKLKMSLPQHLPSLQKLFINDC-------KMLEASIPNGDNIIDLDIK 700
FP L+ L I+ CP+L + +P SL++L ++DC ++ + + I ++DI
Sbjct: 859 FPTLENLSIKNCPELSLEIPIQFSSLKRLEVSDCPVVFDDAQLFRSQLEAMKQIEEIDIC 918
Query: 701 RCDRIL---VNELPTSLKKLFI-------LENRYTEFSVEQIFVNS-------------- 736
C+ + + LPT+LK++ I LE E VE + VN
Sbjct: 919 DCNSVTSFPFSILPTTLKRIQISRCPKLKLEAPVGEMFVEYLRVNDCGCVDDISPEFLPT 978
Query: 737 ------------------TILEVLELDLNGSLKCPTLDLCCYNSLGELSITRWCSSSLSF 778
T E L ++ L + C + S+ W L
Sbjct: 979 ARQLSIENCQNVTRFLIPTATETLR--ISNCENVEKLSVACGGAAQMTSLNIWGCKKLKC 1036
Query: 779 SLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVC 838
L +L L DCP + EG LP NL L I C KL+ R+EW L+ L ++
Sbjct: 1037 LPELLPSLKELRLSDCPEI----EGELPFNLEILRIIYCKKLVNGRKEWHLQRLTELWID 1092
Query: 839 DD--FENVESFPKESLLP-------PTLSYLNLNNCSKLRIMNNEGFL------------ 877
D E++E + + TLS +L + + L+ + EG+L
Sbjct: 1093 HDGSDEDIEHWELPCSIQRLTIKNLKTLSSQHLKSLTSLQYLCIEGYLSQIQSQGQLSSF 1152
Query: 878 -HLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQ 921
HL SL+ L I N +L+ L E ALP+SL L I DCP ++ ++
Sbjct: 1153 SHLTSLQTLQIWNFLNLQSLAESALPSSLSHLEIDDCPNLQSLFE 1197
Score = 68.6 bits (166), Expect = 9e-10
Identities = 100/381 (26%), Positives = 156/381 (40%), Gaps = 73/381 (19%)
Query: 574 LVSLNLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSS---SIN-------- 622
L + + C L P+G + F++ L ++DC + I EF ++ SI
Sbjct: 935 LKRIQISRCPKLKLEAPVGEM-FVEYLRVNDCGCVDDISPEFLPTARQLSIENCQNVTRF 993
Query: 623 VLFRSLEVLKFEKMNNWEEW-LCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDC 681
++ + E L+ N E+ + G + L I C KLK LP+ LPSL++L ++DC
Sbjct: 994 LIPTATETLRISNCENVEKLSVACGGAAQMTSLNIWGCKKLKC-LPELLPSLKELRLSDC 1052
Query: 682 KMLEASIPNGDNIIDLDIKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEV 741
+E +P N+ L I C +++ L++L L + + S E I +
Sbjct: 1053 PEIEGELPF--NLEILRIIYCKKLVNGRKEWHLQRLTELWIDH-DGSDEDIEHWELPCSI 1109
Query: 742 LELDLNGSLKCPTLDLCCYNSLGELSITRWCSS-----SLSFSLHLFTNLYSLWFVDCPN 796
L + + L SL L I + S LS HL T+L +L + N
Sbjct: 1110 QRLTIKNLKTLSSQHLKSLTSLQYLCIEGYLSQIQSQGQLSSFSHL-TSLQTLQIWNFLN 1168
Query: 797 LDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVCDDFENVESFPKESLLPPT 856
L S E LP +L L I +CP L+SL ES LP +
Sbjct: 1169 LQSLAESALPSSLSHLEIDDCP---------NLQSL----------------FESALPSS 1203
Query: 857 LSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLI 916
LS L + +C +L+SL F + +P+SL L I +CPL+
Sbjct: 1204 LSQLFIQDCP-----------NLQSLPF--------------KGMPSSLSKLSIFNCPLL 1238
Query: 917 KVKYQKEGGEQRDTICHIPCV 937
+ + GE I HIP +
Sbjct: 1239 TPLLEFDKGEYWPQIAHIPII 1259
>UniRef100_Q6L3K6 Putative resistance complex protein I2C-2 [Solanum demissum]
Length = 1284
Score = 514 bits (1323), Expect = e-144
Identities = 337/978 (34%), Positives = 516/978 (52%), Gaps = 96/978 (9%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSADGEDLNLLQHQLQYMLMGKKY 60
YND K+K HF KAW+ VSE +D++ +TK +L+ F D +LN LQ +L+ L GKK+
Sbjct: 220 YNDEKVKNHFGFKAWICVSEPYDILRITKELLQEFGLMVDN-NLNQLQVKLKEGLKGKKF 278
Query: 61 LLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKSN 120
L+VLDD+WN + + W+ L F G GSKI+VTTR++ VA ++ ++ L
Sbjct: 279 LIVLDDVWNENYKEWDDLRNLFVQGDVGSKIIVTTRKESVA--LMMGCGAINVGILSSEV 336
Query: 121 CWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILE 180
W+LF H+F+ + EY + VG++I KC GLPLA+K+L +LR F +EW +IL
Sbjct: 337 SWALFKRHSFENRDPEEYSEFQEVGKQIANKCKGLPLALKTLAGILRSKFEVNEWRDILR 396
Query: 181 TDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLLK 240
+++W L + + + L LSY++L +LK+CF++C+I+PK H F K+++I LW+A GL++
Sbjct: 397 SEIWELPRHSNGILPALMLSYNDLRPHLKQCFAFCAIYPKDHLFSKEQVIHLWIANGLVQ 456
Query: 241 CCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQI 300
S N+ F +L S S F++ + ++MHDLVNDL + S I++
Sbjct: 457 QLHS-------ANQYFLELRSRSLFEKVRESSKWNQGEFLMHDLVNDLAQIASSNLCIRL 509
Query: 301 EDARVERSVERTRHIWFSLQSNSVDKLLELT-CEGLHSLILEGT--RAMLISNNVQQDLF 357
E+ + +E+TRH+ +S+ KL L E L +L+ R +S V D+
Sbjct: 510 EENQGSHMLEQTRHLSYSMGDGDFGKLKTLNKLEQLRTLLPINIQLRWCHLSKRVLHDIL 569
Query: 358 SRLNFLRMLSFRGCGLLELVDEIS-NLKLLRYLDLSYTWIEILPDTICMLHNLQTLLLEG 416
RL LR LS E +++ LK LR+LD S+T I+ LPD+IC+L+NL+TLLL
Sbjct: 570 PRLTSLRALSLSHYKNEEFPNDLFIKLKHLRFLDFSWTNIKNLPDSICVLYNLETLLLSY 629
Query: 417 CCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVEEQNVS 474
C L ELP + KL+NLRHL + T P H KL +L L + F++ ++ S
Sbjct: 630 CSNLMELPLHMEKLINLRHLDISE-----AYLTTPLHLSKLKSLDVLVGAKFLLSGRSGS 684
Query: 475 DLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDESMAE 534
+++L KL++L+G++ I GL +V D +S N+++ K++E L +++ G D S E
Sbjct: 685 RMEDLGKLHNLYGSLSILGLQHVVDRRESLKANMREKKHVERLSLEWSGSNA--DNSQTE 742
Query: 535 SNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTL 594
++ L+ LQPN N+K + I+ Y+G FPNW+ + L ++L++C C LP LG L
Sbjct: 743 RDI--LDELQPNTNIKEVEINGYRGTKFPNWLADHSFHKLTKVSLRYCKDCDSLPALGQL 800
Query: 595 PFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEG--FPLLK 652
P LK L+I I + EEFY SSS F SLE L+F +M W++W L FP+L+
Sbjct: 801 PCLKFLTIRGMHQITEVTEEFYGSSSFTKPFNSLEELEFGEMPEWKQWHVLGKGEFPVLE 860
Query: 653 ELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIP----------------------- 689
EL I +CPKL LP++L SL +L I+ C L P
Sbjct: 861 ELSIEDCPKLIGKLPENLSSLTRLRISKCPELSLETPIQLSNLKEFEVANSPKVGVVFDD 920
Query: 690 ---------NGDNIIDLDIKRCDRIL---VNELPTSLKKLFILENRYTEFS--------V 729
I+ LDI C + ++ LP++LK++ I R + V
Sbjct: 921 AQLFTSQLEGMKQIVKLDITDCKSLTSLPISILPSTLKRIRISGCRELKLEAPINAICRV 980
Query: 730 EQIFVNSTILEVLELDLNGSLKCPT---------------LDLCCYNSLGELSITRWCSS 774
+ + L V + L PT L + C + L I C
Sbjct: 981 PEFLPRALSLSVRSCNNLTRLLIPTATETVSIRDCDNLEILSVACGTQMTSLHIYH-CEK 1039
Query: 775 SLSFSLH---LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKS 831
S H L +L L V+C ++SFPEGGLP NL L I+ C KL+ R+EW L+
Sbjct: 1040 LKSLPEHMQQLLPSLKELKLVNCSQIESFPEGGLPFNLQQLWISCCKKLVNGRKEWHLQR 1099
Query: 832 LKYF----FVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYI 887
L D + V ++ LP ++ L++ N L+ ++++ L SLE+L+
Sbjct: 1100 LPCLRDLTIHHDGSDEVVLADEKWELPCSIRRLSIWN---LKTLSSQLLKSLTSLEYLFA 1156
Query: 888 INCPSLERLPEEALPNSL 905
N P ++ L EE LP+SL
Sbjct: 1157 NNLPQMQSLLEEGLPSSL 1174
Score = 42.7 bits (99), Expect = 0.051
Identities = 106/449 (23%), Positives = 171/449 (37%), Gaps = 102/449 (22%)
Query: 548 NLKRLTISKYKGNSFPN---------------WIRGYHLP-------NLVSLNLQFCGLC 585
+L+ L++S YK FPN W +LP NL +L L +C
Sbjct: 574 SLRALSLSHYKNEEFPNDLFIKLKHLRFLDFSWTNIKNLPDSICVLYNLETLLLSYCSNL 633
Query: 586 SLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEK-MNNWEEWLC 644
LP L K++++ D I E + + +SL+VL K + +
Sbjct: 634 MELP----LHMEKLINLRHLD----ISEAYLTTPLHLSKLKSLDVLVGAKFLLSGRSGSR 685
Query: 645 LEGFPLLKELY-------------IRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNG 691
+E L LY RE K M +H+ L LE S N
Sbjct: 686 MEDLGKLHNLYGSLSILGLQHVVDRRESLKANMREKKHVERLS---------LEWSGSNA 736
Query: 692 DNIIDLDIKRCDRILVNELP--TSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGS 749
DN + +R +++EL T++K++ I R T+F + + ++ ++ L
Sbjct: 737 DN------SQTERDILDELQPNTNIKEVEINGYRGTKFP--NWLADHSFHKLTKVSLRYC 788
Query: 750 LKC---------PTLDLCCYNSLGELS-ITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDS 799
C P L + +++ +T S SF+ F +L L F + P
Sbjct: 789 KDCDSLPALGQLPCLKFLTIRGMHQITEVTEEFYGSSSFTKP-FNSLEELEFGEMPEWKQ 847
Query: 800 FP---EGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVCDDFENVESFPKESLLPP- 855
+ +G P L L+I +CPKLI E L SL + P+ SL P
Sbjct: 848 WHVLGKGEFPV-LEELSIEDCPKLIGKLPE-NLSSLTRL-------RISKCPELSLETPI 898
Query: 856 ---TLSYLNLNNCSKLRIMNNEGFLHLKSLEF------LYIINCPSLERLPEEALPNSLY 906
L + N K+ ++ ++ L LE L I +C SL LP LP++L
Sbjct: 899 QLSNLKEFEVANSPKVGVVFDDAQLFTSQLEGMKQIVKLDITDCKSLTSLPISILPSTLK 958
Query: 907 SLWIKDCPLIKVKYQKEGGEQRDTICHIP 935
+ I C +K++ + IC +P
Sbjct: 959 RIRISGCRELKLE------APINAICRVP 981
>UniRef100_Q6L3G4 Putative plant disease resistant protein [Solanum demissum]
Length = 1406
Score = 497 bits (1279), Expect = e-139
Identities = 367/1104 (33%), Positives = 537/1104 (48%), Gaps = 185/1104 (16%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILK---SFNSSADGEDLNLLQHQLQYMLMG 57
YND K+K HF LKAW VSE +D + +TK +L+ SF+S AD +LN LQ +L+ +L G
Sbjct: 314 YNDEKVKNHFNLKAWFCVSEPYDALRITKGLLQEIGSFDSKADS-NLNQLQVKLKEILKG 372
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
K++L+VLDD+WN + W+ L F G GSKI+VTTR++ VA ++ E ++ L
Sbjct: 373 KRFLIVLDDMWNDNYNEWDDLRNLFVKGDVGSKIIVTTRKESVA--LVMGKEQISMEILS 430
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
WSLF HAF+ E L+ VG++IV KC GLPLA+K+L +LR W
Sbjct: 431 SEVSWSLFKRHAFEYMDPEEQRELKKVGKQIVAKCKGLPLALKTLAGMLRSKSEVEGWKR 490
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
IL ++MW L D+++ L LSY++LP++LK+CFSYC+IFPK + F+K+++I LW+A G
Sbjct: 491 ILRSEMWELP--DNDILPALMLSYNDLPTHLKQCFSYCAIFPKDYPFRKEQVIQLWIANG 548
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFS 297
LLK + + E+ GN F +L S S F++ + E ++MHDL+NDL + S +
Sbjct: 549 LLKGLQKDETIEDLGNLYFLELRSRSLFERVRESSKRNEEEFLMHDLINDLAQVASSKLC 608
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSVDKLLEL-TCEGLHSLI---LEGTRAMLISNNVQ 353
I++ED +E+ R++ +SL +KL L + L +L+ ++ + +S V
Sbjct: 609 IRLEDNEGSHMLEKCRNLSYSLGDGVFEKLKPLYKSKQLRTLLPINIQRGYSFPLSKRVL 668
Query: 354 QDLFSRLNFLRMLSFRGCGLLELVDEI-SNLKLLRYLDLSYTWIEILPDTICMLHNLQTL 412
++ RL LR LS + EL +++ LKLLR LDLS T I LPD+IC L+NL+ L
Sbjct: 669 YNILPRLTSLRALSLSHYRIKELPNDLFITLKLLRILDLSQTAIRKLPDSICALYNLEIL 728
Query: 413 LLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVEEQN 472
LL C L ELP + KL+NLRHL G +K MP H KL NL L F
Sbjct: 729 LLSSCIYLEELPPHMEKLINLRHL---DTTGTSLLK-MPLHPSKLKNLHVLVGFKFILGG 784
Query: 473 VSDLK--ELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDE 530
+DL+ +L +L++LHG+I + L NV D ++ N+ +++E L + E E
Sbjct: 785 CNDLRMVDLGELHNLHGSISVLELQNVVDRREALNANMMKKEHVEMLSL-------EWSE 837
Query: 531 SMAESNVS---VLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSL 587
S+A+S+ + +L+ LQPN N+K L I+ Y+G FPNW+ + LV ++L C C+
Sbjct: 838 SIADSSQTEGDILDKLQPNTNIKELEIAGYRGTKFPNWMADHSFLKLVGVSLSNCNNCAS 897
Query: 588 LPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCL-- 645
LP LG LP LK L++ I + EEFY + S F SLE L+F +M W++W L
Sbjct: 898 LPALGQLPSLKFLTVRGMHRITEVSEEFYGTLSSKKPFNSLEKLEFAEMPEWKQWHVLGK 957
Query: 646 EGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIP---------------- 689
FP L + I +CPKL LP+ L SL+ L I+ C L P
Sbjct: 958 GEFPALHDFLIEDCPKLIGKLPEKLCSLRGLRISKCPELSPETPIQLSNLKEFKVVASPK 1017
Query: 690 ----------------NGDNIIDLDIKRCDRIL---VNELPTSLKKLFILENRYTEFSVE 730
I++L I C + ++ LP++LKK+ I R +
Sbjct: 1018 VGVLFDDAQLFTSQLQGMKQIVELCIHDCHSLTFLPISILPSTLKKIEIYHCRKLKLEAS 1077
Query: 731 QI-------FVNSTILEVLEL--DLNGSL----------KCPTLDLCCYNSLGELSITRW 771
I F+ + ++ + D++ L CP L + E
Sbjct: 1078 MISRGDCNMFLENLVIYGCDSIDDISPELVPRSHYLSVNSCPNLTRLLIPTETEKLYIWH 1137
Query: 772 CSS--SLSFSLHLFTNLYSLWFVDCPNLD-------------------------SFPEGG 804
C + LS + T L +L DC L SFPEGG
Sbjct: 1138 CKNLEILSVASGTQTMLRNLSIRDCEKLKWLPECMQELIPSLKELELWFCTEIVSFPEGG 1197
Query: 805 LPCNLLSLTITNCPKLIASRQEW------------------------------------- 827
LP NL L I C KL+ +R+EW
Sbjct: 1198 LPFNLQVLRIHYCKKLVNARKEWHLQRLPCLRELTILHDGSDLAGENWELPCSIRRLTVS 1257
Query: 828 -----------GLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGF 876
L SL+Y + + ++S +E LP +LS L L +L + EG
Sbjct: 1258 NLKTLSSQLFKSLTSLEYLSTGNSLQ-IQSLLEEG-LPISLSRLTLFGNHELHSLPIEGL 1315
Query: 877 LHLKSLEFLYIINCPSLERLPEEALPNSL-----------------------YSLWIKDC 913
L SL L+I +C L+ +PE ALP+SL SL I DC
Sbjct: 1316 RQLTSLRDLFISSCDQLQSVPESALPSSLSELTIQNCHKLQYLPVKGMPTSISSLSIYDC 1375
Query: 914 PLIKVKYQKEGGEQRDTICHIPCV 937
PL+K + + GE I HI +
Sbjct: 1376 PLLKPLLEFDKGEYWPKIAHISTI 1399
>UniRef100_Q6L3G6 Putative plant disease resistant protein [Solanum demissum]
Length = 1315
Score = 496 bits (1277), Expect = e-138
Identities = 339/1009 (33%), Positives = 526/1009 (51%), Gaps = 117/1009 (11%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSS--ADGEDLNLLQHQLQYMLMGK 58
YND K+K+HF LKAW+ VSE +D V +TK +L+ +SS +LN LQ +L+ L GK
Sbjct: 206 YNDEKVKDHFGLKAWICVSEPYDAVRITKELLQEISSSDCTGNSNLNQLQIKLKESLKGK 265
Query: 59 KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDK 118
K+L+VLDD+WN + + W+ L F G GSKI+VTTR++ VA ++ +L L
Sbjct: 266 KFLIVLDDVWNENYDEWDDLRNIFVQGDIGSKIIVTTRKESVA--LMMGCGAVNLGTLSS 323
Query: 119 SNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINI 178
W+LF H+ + + E+P LE VG++I KC GLPLA+K+L +LR +EW +I
Sbjct: 324 EVSWALFKRHSLENRGPEEHPELEEVGKQIAHKCKGLPLALKALAGILRSKSDLNEWRDI 383
Query: 179 LETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGL 238
L +++W L + + L LSY++LP++LKRCF++C+I+PK + F K+++I LW+A GL
Sbjct: 384 LRSEIWELPSHSNGILPALMLSYNDLPAHLKRCFAFCAIYPKDYMFCKEQVIHLWIANGL 443
Query: 239 LKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSI 298
+ + + GN+ F +L S S F++ + E ++MHDLVNDL + S I
Sbjct: 444 V-------PQLDSGNQYFLELRSRSLFERIPESSKWNSEEFLMHDLVNDLAQIASSNLCI 496
Query: 299 QIEDARVERSVERTRHIWFSLQSNSVDKLLEL-TCEGLHSLI---LEGTRAMLISNNVQQ 354
++E+ + +E++RHI +S +KL L E L +L+ ++ +S V
Sbjct: 497 RLEENQGSHMLEQSRHISYSTGEGDFEKLKPLFKSEQLRTLLPISIQRDYLFKLSKRVLH 556
Query: 355 DLFSRLNFLRMLSFRGCGLLELVDEI-SNLKLLRYLDLSYTWIEILPDTICMLHNLQTLL 413
++ RL LR LS ++EL +++ LKLLR+LD+S T I+ LPD+IC+L+NL+ LL
Sbjct: 557 NVLPRLTSLRALSLSPYKIVELPNDLFIKLKLLRFLDISRTKIKKLPDSICVLYNLEILL 616
Query: 414 LEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVEEQ 471
L C +L ELP KL+NL +L + N +K MP H KL +L L + F++ +
Sbjct: 617 LSSCDDLEELPLQMEKLINLHYLDI---NNTSRLK-MPLHLSKLKSLHVLVGAKFLLGGR 672
Query: 472 NVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDES 531
S + +L ++++L G++ I L NV D ++ N+K+ ++E L +++ R D S
Sbjct: 673 GGSRMDDLGEVHNLFGSLSILELQNVVDRWEALKANMKEKNHVEMLSLEW--SRSIADNS 730
Query: 532 MAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPL 591
E + +L+ LQPN N+ L I Y+G FPNW+ LV L+L C C LP L
Sbjct: 731 KNEKD--ILDGLQPNTNINELQIGGYRGTKFPNWLADQSFLKLVQLSLSNCKDCDSLPAL 788
Query: 592 GTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEG--FP 649
G LP LK L+I I + EEFY S S F SLE L+F +M W+ W L FP
Sbjct: 789 GQLPSLKFLAIRRMRRIIEVTEEFYGSLSSKKPFNSLEKLEFAEMPEWKRWHVLGNGEFP 848
Query: 650 LLKELYIRECPKLKMSLPQHLPSLQKLFINDC---------------------------- 681
LK L + +CPKL P++L SL L I+ C
Sbjct: 849 ALKILSVEDCPKLIEKFPENLSSLTGLRISKCPELSLETSIQLSTLKIFEVISSPKVGVL 908
Query: 682 ----KMLEASIPNGDNIIDLDIKRCDRIL---VNELPTSLKKLFI--LENRYTEFSVEQI 732
++ + + +I++L C+ + ++ LP++LK++ I E + V ++
Sbjct: 909 FDDTELFTSQLQEMKHIVELFFTDCNSLTSLPISILPSTLKRIHIYQCEKLKLKTPVGEM 968
Query: 733 FVNSTILEVLELDLNGSL---------KCPTLDLCCYNSLGEL-------SITRWCSSSL 776
N+ LE L+LD S+ + TL + +SL L S+T W +L
Sbjct: 969 ITNNMFLEELKLDGCDSIDDISPELVPRVGTLIVGRCHSLTRLLIPTETKSLTIWSCENL 1028
Query: 777 SF-----------------------------SLHLFTNLYSLWFVDCPNLDSFPEGGLPC 807
L +L +L +CP + SFPEGGLP
Sbjct: 1029 EILSVACGARMMSLRFLNIENCEKLKWLPECMQELLPSLNTLELFNCPEMMSFPEGGLPF 1088
Query: 808 NLLSLTITNCPKLIASRQEWGLK---SLKYFFVCDDFENVESFPKESL-LPPTLSYLNLN 863
NL L I NC KL+ R+ W L+ L+ + D + E E+ LP ++ L ++
Sbjct: 1089 NLQVLLIWNCKKLVNGRKNWRLQRLPCLRELRIEHDGSDEEILAGENWELPCSIQRLYIS 1148
Query: 864 NCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKD 912
N L+ ++++ L SL +L P ++ L EE LP+SLY L + D
Sbjct: 1149 N---LKTLSSQVLKSLTSLAYLDTYYLPQIQSLLEEGLPSSLYELRLDD 1194
Score = 53.9 bits (128), Expect = 2e-05
Identities = 131/543 (24%), Positives = 207/543 (37%), Gaps = 76/543 (13%)
Query: 335 LHSLILEGTRAMLISNNVQQDLFSRLNFLRMLSFRGCGLLELVDEISNLKLLRYLDLSYT 394
++ L + G R N + F +L L + + + C L + ++ +LK L +
Sbjct: 746 INELQIGGYRGTKFPNWLADQSFLKLVQLSLSNCKDCDSLPALGQLPSLKFLAIRRMRRI 805
Query: 395 WIEILPDTICMLHNLQTLLLEGCCELTELPS--NFSKLVNLRHLKLPSHNGRPCIKTMPK 452
IE+ + L + + E E+P + L N L + C K + K
Sbjct: 806 -IEVTEEFYGSLSSKKPFNSLEKLEFAEMPEWKRWHVLGNGEFPALKILSVEDCPKLIEK 864
Query: 453 HTGKLNNLQSLSYFIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTK 512
L++L L E ++ +L+ L + I +G + D + T L++ K
Sbjct: 865 FPENLSSLTGLRISKCPELSLETSIQLSTLK-IFEVISSPKVGVLFDDTELFTSQLQEMK 923
Query: 513 YLEELHMKFDGGREEMDESMAESNVSVLEALQPNR------------NLKRLTISKYKGN 560
++ EL + S+ S + + Q + N L K G
Sbjct: 924 HIVELFFTDCNSLTSLPISILPSTLKRIHIYQCEKLKLKTPVGEMITNNMFLEELKLDGC 983
Query: 561 SFPNWIRGYHLPNLVSLNLQFC-GLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSS 619
+ I +P + +L + C L LL P T K L+I C+ ++I+ S
Sbjct: 984 DSIDDISPELVPRVGTLIVGRCHSLTRLLIPTET----KSLTIWSCENLEIL------SV 1033
Query: 620 SINVLFRSLEVLKFEKMNNWEEWL--CL-EGFPLLKELYIRECPKLKMSLPQH-LP-SLQ 674
+ SL L E + WL C+ E P L L + CP++ MS P+ LP +LQ
Sbjct: 1034 ACGARMMSLRFLNIENCEKLK-WLPECMQELLPSLNTLELFNCPEM-MSFPEGGLPFNLQ 1091
Query: 675 KLFINDCKMLEASIPNGDNIIDLDIKRC------------DRILVNE---LPTSLKKLFI 719
L I +CK L NG L C + IL E LP S+++L+I
Sbjct: 1092 VLLIWNCKKLV----NGRKNWRLQRLPCLRELRIEHDGSDEEILAGENWELPCSIQRLYI 1147
Query: 720 LENRYTEFSVEQIFVNSTILE---------VLELDLNGSL------------KCPTLDLC 758
+ V + + L+ +LE L SL PT L
Sbjct: 1148 SNLKTLSSQVLKSLTSLAYLDTYYLPQIQSLLEEGLPSSLYELRLDDHHELHSLPTKGLR 1207
Query: 759 CYNSLGELSITRWCSSSLSFSLH-LFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNC 817
SL L I R C+ S + L +++ L CPNL S P G+P +L L I NC
Sbjct: 1208 HLTSLRRLEI-RHCNQLQSLAESTLPSSVSELTIGYCPNLQSLPVKGMPSSLSKLHIYNC 1266
Query: 818 PKL 820
P L
Sbjct: 1267 PLL 1269
Score = 37.4 bits (85), Expect = 2.2
Identities = 45/141 (31%), Positives = 61/141 (42%), Gaps = 29/141 (20%)
Query: 783 FTNLYSLWFVDCPNL-DSFPEGGLPCNLLSLT---ITNCPKLIASRQEWGLKSLKYFFVC 838
F L L DCP L + FPE NL SLT I+ CP+L + L +LK F
Sbjct: 847 FPALKILSVEDCPKLIEKFPE-----NLSSLTGLRISKCPEL-SLETSIQLSTLKIF--- 897
Query: 839 DDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPE 898
V S PK +L S+L+ M K + L+ +C SL LP
Sbjct: 898 ----EVISSPKVGVLFDDTELFT----SQLQEM--------KHIVELFFTDCNSLTSLPI 941
Query: 899 EALPNSLYSLWIKDCPLIKVK 919
LP++L + I C +K+K
Sbjct: 942 SILPSTLKRIHIYQCEKLKLK 962
>UniRef100_Q60D49 Putative disease resistance protein I2C-5 [Solanum demissum]
Length = 1266
Score = 496 bits (1277), Expect = e-138
Identities = 340/1018 (33%), Positives = 532/1018 (51%), Gaps = 124/1018 (12%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKS---FNSSADGEDLNLLQHQLQYMLMG 57
YN+ ++K HF LKAW VSE +D + +TK +L+ F+S +LN LQ +L+ L G
Sbjct: 222 YNNERVKNHFGLKAWYCVSEPYDALRITKGLLQEIGKFDSKDVHNNLNQLQVKLKESLKG 281
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
KK+L+VLDD+WN + +W L F G GSKI+VTTR++ VA ++ + + L
Sbjct: 282 KKFLIVLDDVWNNNYNKWVELKNVFVQGDIGSKIIVTTRKESVA--LMMGNKKVSMDNLS 339
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
WSLF HAF+ +P LE VG++I +KC GLPLA+K+L +LR EW
Sbjct: 340 TEASWSLFKRHAFENMDPMGHPELEEVGKQIADKCKGLPLALKTLAGMLRSKSEVEEWKR 399
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
IL +++W L D+++ L LSY++LP +LKRCFSYC+IFPK + F+K+++I LW+A G
Sbjct: 400 ILRSEIWELP--DNDILPALMLSYNDLPVHLKRCFSYCAIFPKDYPFRKEQVIHLWIANG 457
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH-YVMHDLVNDLTKSVSGEF 296
++ ++ ++ GN+ F +L S S F++ + E ++MHDLVNDL + S +
Sbjct: 458 IVP--KDDQIIQDSGNQYFLELRSRSLFEKVPNPSKRNIEELFLMHDLVNDLAQIASSKL 515
Query: 297 SIQIEDARVERSVERTRHIWFSL-QSNSVDKLLEL-TCEGLHSLILEGTRAML-----IS 349
I++E+++ +E++RH+ +S+ + +KL L E L +L+ + +S
Sbjct: 516 CIRLEESKGSDMLEKSRHLSYSMGRGGDFEKLTPLYKLEQLRTLLPTCISTVNYCYHPLS 575
Query: 350 NNVQQDLFSRLNFLRMLSFRGCGLLELVDEIS-NLKLLRYLDLSYTWIEILPDTICMLHN 408
V + RL LR+LS + EL +++ LKLLR+LD+S T I+ LPD+IC+L+N
Sbjct: 576 KRVLHTILPRLRSLRVLSLSHYNIKELPNDLFIKLKLLRFLDISQTEIKRLPDSICVLYN 635
Query: 409 LQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYF 466
L+ LLL C L ELP KL+NL HL + + + + MP H KL +LQ L + F
Sbjct: 636 LEILLLSSCDYLEELPLQMEKLINLHHLDISNTH----LLKMPLHLSKLKSLQVLVGAKF 691
Query: 467 IVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGRE 526
++ + DL E +L+G++ + L NV D ++ +++ +++ L +++ E
Sbjct: 692 LLSGWGMEDLGEA---QNLYGSLSVVELQNVVDRREAVKAKMREKNHVDMLSLEWS---E 745
Query: 527 EMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCS 586
+++ +L+ L P++N+K + I+ Y+G FPNW+ LV L++ C CS
Sbjct: 746 SSSADNSQTERDILDELSPHKNIKEVKITGYRGTKFPNWLADPLFLKLVQLSVVNCKNCS 805
Query: 587 LLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLE 646
LP LG LP LK LSIS GI + EEFY S S F SL L+FE M W++W L
Sbjct: 806 SLPSLGQLPCLKFLSISGMHGITELSEEFYGSLSSKKPFNSLVELRFEDMPKWKQWHVLG 865
Query: 647 G--FPLLKELYIRECPKLKMSLPQHLPSLQKLF--------INDCKMLEASIPNGDNIID 696
F L++L I+ CP+L + P L L K+F D ++ + + I++
Sbjct: 866 SGEFATLEKLLIKNCPELSLETPIQLSCL-KMFEVIGCPKVFGDAQVFRSQLEGTKQIVE 924
Query: 697 LDIKRCDRIL---VNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLEL-------DL 746
LDI C+ + + LPT+LK + I + + V V LE L L D+
Sbjct: 925 LDISDCNSVTSFPFSILPTTLKTITIFGCQKLKLEVP---VGEMFLEYLSLKECDCIDDI 981
Query: 747 NGSL------------------------------KCPTLDL----CCYNSLGELSI---- 768
+ L C +++ C + L+I
Sbjct: 982 SPELLPTARTLYVSNCHNLTRFLIPTATESLYIHNCENVEILSVVCGGTQMTSLTIYMCK 1041
Query: 769 -TRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEW 827
+W + L +L L+ ++CP ++SFPEGGLP NL L I NC KL+ R+EW
Sbjct: 1042 KLKWLPERMQ---ELLPSLKHLYLINCPEIESFPEGGLPFNLQFLQIYNCKKLVNGRKEW 1098
Query: 828 GLKSLKYFFVC--------------DDFE-----------NVESFPKESLLPPT-LSYLN 861
L+ L V +++E N+++ + L T L YL
Sbjct: 1099 RLQRLPCLNVLVIEHDGSDEEIVGGENWELPSSIQRLTIYNLKTLSSQVLKSLTSLQYLC 1158
Query: 862 L-NNCSKLRIMNNEG-FLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIK 917
+ N +++ M +G F HL SL+ L I N P+L+ LPE ALP+SL L I CP ++
Sbjct: 1159 IEGNLPQIQSMLEQGQFSHLTSLQSLEIRNFPNLQSLPESALPSSLSQLTIVYCPKLQ 1216
Score = 51.6 bits (122), Expect = 1e-04
Identities = 90/332 (27%), Positives = 133/332 (39%), Gaps = 63/332 (18%)
Query: 543 LQPNRNLKRLTISKYKG-NSFPNWIRGYHLPN-LVSLNLQFCGLCSLLPPLGTLPFLKML 600
L+ + + L IS SFP I LP L ++ + C L P+G + FL+ L
Sbjct: 916 LEGTKQIVELDISDCNSVTSFPFSI----LPTTLKTITIFGCQKLKLEVPVGEM-FLEYL 970
Query: 601 SISDCDGIKIIGEEFYDS------SSINVLFRSL-----EVLKFEKMNNWEEWLCLEGFP 649
S+ +CD I I E + S+ + L R L E L N E + G
Sbjct: 971 SLKECDCIDDISPELLPTARTLYVSNCHNLTRFLIPTATESLYIHNCENVEILSVVCGGT 1030
Query: 650 LLKELYIRECPKLKMSLPQH----LPSLQKLFINDCKMLEASIPNGD---NIIDLDIKRC 702
+ L I C KLK LP+ LPSL+ L++ +C +E S P G N+ L I C
Sbjct: 1031 QMTSLTIYMCKKLKW-LPERMQELLPSLKHLYLINCPEIE-SFPEGGLPFNLQFLQIYNC 1088
Query: 703 DRILVNELPTSLKKLFIL-----------------ENRYTEFSVEQIFVNSTILEVLELD 745
+++ L++L L EN S++++ + + L+ L
Sbjct: 1089 KKLVNGRKEWRLQRLPCLNVLVIEHDGSDEEIVGGENWELPSSIQRLTIYN--LKTLSSQ 1146
Query: 746 LNGSLK-----CPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYSL----------- 789
+ SL C +L S+ E +S S + F NL SL
Sbjct: 1147 VLKSLTSLQYLCIEGNLPQIQSMLEQGQFSHLTSLQSLEIRNFPNLQSLPESALPSSLSQ 1206
Query: 790 -WFVDCPNLDSFPEGGLPCNLLSLTITNCPKL 820
V CP L S P G+P +L L+I CP L
Sbjct: 1207 LTIVYCPKLQSLPVKGMPSSLSELSIYQCPLL 1238
>UniRef100_O24015 Resistance complex protein I2C-1 [Lycopersicon esculentum]
Length = 1220
Score = 482 bits (1240), Expect = e-134
Identities = 337/1034 (32%), Positives = 529/1034 (50%), Gaps = 131/1034 (12%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSS---ADG------------EDLN 45
YND ++++HF L AW VSE++D +TK +L+ S+ AD ++LN
Sbjct: 214 YNDERVQKHFGLTAWFCVSEAYDAFRITKGLLQEIGSTDLKADDNLNQLQVKLKADDNLN 273
Query: 46 LLQHQLQYMLMGKKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVL 105
LQ +L+ L GK++L+VLDD+WN + W+ L F G GSKI+VTTR++ VA ++
Sbjct: 274 QLQVKLKEKLNGKRFLVVLDDVWNDNYPEWDDLRNLFLQGDIGSKIIVTTRKESVA--LM 331
Query: 106 KSTELFDLQQLDKSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQL 165
+ + L + W+LF H+ + K E+P E VG++I +KC GLPLA+K+L +
Sbjct: 332 MDSGAIYMGILSSEDSWALFKRHSLEHKDPKEHPEFEEVGKQIADKCKGLPLALKALAGM 391
Query: 166 LRKTFSEHEWINILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFK 225
LR EW NIL +++W L + + L LSY++LP++LK+CF+YC+I+PK ++F+
Sbjct: 392 LRSKSEVDEWRNILRSEIWELPSCSNGILPALMLSYNDLPAHLKQCFAYCAIYPKDYQFR 451
Query: 226 KDELIMLWMAEGLLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLV 285
K+++I LW+A GL+ + GN+ F +L S S F+ + + E ++MHDLV
Sbjct: 452 KEQVIHLWIANGLV-------HQFHSGNQYFIELRSRSLFEMASEPSERDVEEFLMHDLV 504
Query: 286 NDLTKSVSGEFSIQIEDARVERSVERTRHIWFSL-QSNSVDKLLEL-TCEGLHSLI---L 340
NDL + S I++ED + +E+ RH+ +S+ Q +KL L E L +L+ +
Sbjct: 505 NDLAQIASSNHCIRLEDNKGSHMLEQCRHMSYSIGQDGEFEKLKSLFKSEQLRTLLPIDI 564
Query: 341 EGTRAMLISNNVQQDLFSRLNFLRMLSFRGCGLLELVDEI-SNLKLLRYLDLSYTWIEIL 399
+ + +S V ++ L LR LS + L +++ LKLLR+LDLS T I L
Sbjct: 565 QFHYSKKLSKRVLHNILPTLRSLRALSLSHYQIEVLPNDLFIKLKLLRFLDLSETSITKL 624
Query: 400 PDTICMLHNLQTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNN 459
PD+I +L+NL+TLLL C L ELP KL+NLRHL + S+ R MP H +L +
Sbjct: 625 PDSIFVLYNLETLLLSSCEYLEELPLQMEKLINLRHLDI-SNTRR---LKMPLHLSRLKS 680
Query: 460 LQSL--SYFIVEEQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEEL 517
LQ L + F+V ++ L + ++L+G++ I L NV D ++ +++ ++E+L
Sbjct: 681 LQVLVGAKFLVGGWR---MEYLGEAHNLYGSLSILELENVVDRREAVKAKMREKNHVEQL 737
Query: 518 HMKFDGGREEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSL 577
+++ E + +++ +L+ L+P++N+K + I+ Y+G +FPNW+ LV L
Sbjct: 738 SLEWS---ESISADNSQTERDILDELRPHKNIKAVEITGYRGTNFPNWVADPLFVKLVHL 794
Query: 578 NLQFCGLCSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMN 637
L+ C C LP LG LP L+ LSI GI+++ EEFY S F SL L+FE M
Sbjct: 795 YLRNCKDCYSLPALGQLPCLEFLSIRGMHGIRVVTEEFYGRLSSKKPFNSLVKLRFEDMP 854
Query: 638 NWEEW--LCLEGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCK------------- 682
W++W L + FP L++L I+ CP+L + +P SL++L I DCK
Sbjct: 855 EWKQWHTLGIGEFPTLEKLSIKNCPELSLEIPIQFSSLKRLDICDCKSVTSFPFSILPTT 914
Query: 683 ----------MLEASIPNGDNIID----LDIKRCDRILVNELPTSLKKLFILENRYTEFS 728
L+ P G+ ++ +D D I LPT+ + +EN +
Sbjct: 915 LKRIKISGCPKLKLEAPVGEMFVEYLSVIDCGCVDDISPEFLPTA--RQLSIENCHNVTR 972
Query: 729 VEQIFVNSTILEVLELDLNGSLKCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYS 788
F+ T E L + C L + C + S+ W L L +L
Sbjct: 973 ----FLIPTATESLHI-----RNCEKLSMACGGAAQLTSLNIWGCKKLKCLPELLPSLKE 1023
Query: 789 LWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVCDD--FENVES 846
L CP + EG LP NL L I C KL+ R+EW L+ L ++ D E++E
Sbjct: 1024 LRLTYCPEI----EGELPFNLQILDIRYCKKLVNGRKEWHLQRLTELWIKHDGSDEHIEH 1079
Query: 847 FPKESLLP-------PTLSYLNLNNCSK---LRIMNN----------EGFLHLKSLE--- 883
+ S + TLS +L + + LRI+ N F HL SL+
Sbjct: 1080 WELPSSIQRLFIFNLKTLSSQHLKSLTSLQFLRIVGNLSQFQSQGQLSSFSHLTSLQTLQ 1139
Query: 884 --------------------FLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKE 923
L I NCP+L+ LP + +P+SL +L I CPL+ + +
Sbjct: 1140 IWNFLNLQSLPESALPSSLSHLIISNCPNLQSLPLKGMPSSLSTLSISKCPLLTPLLEFD 1199
Query: 924 GGEQRDTICHIPCV 937
GE I HIP +
Sbjct: 1200 KGEYWTEIAHIPTI 1213
>UniRef100_Q945S6 I2C-5 [Lycopersicon pimpinellifolium]
Length = 1297
Score = 477 bits (1228), Expect = e-133
Identities = 344/1024 (33%), Positives = 525/1024 (50%), Gaps = 134/1024 (13%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKS---FNSSADGEDLNLLQHQLQYMLMG 57
YND ++++HF LKAW VSE +D +TK +L+ F+S +LN LQ +L+ L G
Sbjct: 222 YNDERVQKHFVLKAWFCVSEVYDAFTITKGLLQEIGKFDSKDVHNNLNQLQVKLKESLKG 281
Query: 58 KKYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
KK+L+VLDD+WN + W L F G GSKI+VTTR+ VA ++ E + L
Sbjct: 282 KKFLIVLDDVWNENYNEWNDLRNIFVQGDIGSKIIVTTRKDSVA--LMMGNEQISMGNLS 339
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
WSLF HAF+ +P LE VGR+I KC GLPLA+K+L +LR EW
Sbjct: 340 TEASWSLFKRHAFENMDPMGHPELEEVGRQIAAKCKGLPLALKTLAGMLRPKSEIDEWKC 399
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
IL +++W L D+++ L LSY++LP++LKRCFS+C+IFPK + F+K+++I LW+A G
Sbjct: 400 ILRSEIWELR--DNDILPALMLSYNDLPAHLKRCFSFCAIFPKDYPFRKEQVIHLWIANG 457
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEH-YVMHDLVNDLTKSVSGEF 296
L+ + ++ GN+ F +L S S F++ + E ++MHDLVNDL + S +
Sbjct: 458 LVPV--KDEINQDLGNQYFLELRSRSLFEKVPNPSKRNIEELFLMHDLVNDLAQIASSKL 515
Query: 297 SIQIEDARVERSVERTRHIWFSL-QSNSVDKLLEL-TCEGLHSL--ILEGTRAMLISNNV 352
I++E+ + +E++ H+ +S+ + +KL L E L +L I R+ +S V
Sbjct: 516 CIRLEERKGSFMLEKSWHVSYSMGRDGEFEKLTPLYKLEQLRTLLPIRIEFRSHYLSKRV 575
Query: 353 QQDLFSRLNFLRMLSFRGCGLLELVDEIS-NLKLLRYLDLSYTWIEILPDTICMLHNLQT 411
++ L LR+LS EL +++ LKLLR+LDLS TWI LPD+IC L+NL+T
Sbjct: 576 LHNILPTLRSLRVLSLSHYKNKELPNDLFIKLKLLRFLDLSCTWITKLPDSICGLYNLET 635
Query: 412 LLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVE 469
LLL C +L ELP KL+NLRHL + S+ R MP H +L +LQ L + F+V
Sbjct: 636 LLLSSCYKLEELPLQMEKLINLRHLDV-SNTRR---LKMPLHLSRLKSLQVLVGAEFLVV 691
Query: 470 EQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMD 529
++ L + +L+G++ + L NV + ++ +++ ++E+L +++ D
Sbjct: 692 GWR---MEYLGEAQNLYGSLSVVKLENVVNRREAVKAKMREKNHVEQLSLEWSKS-SIAD 747
Query: 530 ESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLP 589
S E ++ L+ L P++N+K + IS Y+G +FPNW+ LV L+L +C C LP
Sbjct: 748 NSQTERDI--LDELHPHKNIKEVVISGYRGTNFPNWVADPLFVKLVKLSLSYCKDCYSLP 805
Query: 590 PLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEW--LCLEG 647
LG LP LK LS+ GI+++ EEFY S F LE LKFE M W++W L +
Sbjct: 806 ALGQLPCLKFLSVKGMHGIRVVTEEFYGRLSSKKPFNCLEKLKFEDMTEWKQWHALGIGE 865
Query: 648 FPLLKELYIRECPKLKMSLPQHLPSLQKLFI-------NDCKMLEASIPNGDNIIDLDIK 700
FP L++L I+ CP+L + P SL++L + +D ++ + I L+I
Sbjct: 866 FPTLEKLSIKNCPELSLERPIQFSSLKRLEVVGCPVVFDDAQLFRFQLEAMKQIEALNIS 925
Query: 701 RCDRIL---VNELPTSLKKLFI-------LENRYTEFSVEQIFVNST----------ILE 740
C+ + + LPT+LK++ I E E VE + V++ I
Sbjct: 926 DCNSVTSFPFSILPTTLKRIQISGCPKLKFEVPVCEMFVEYLGVSNCDCVDDMSPEFIPT 985
Query: 741 VLELDLNGS-------LKCPTLDLCCYNSLGELSITRWCS-----SSLSFS--------- 779
+L + + T LC +N ++ C +SL+ S
Sbjct: 986 ARKLSIESCHNVTRFLIPTATETLCIFNCENVEKLSVACGGAAQLTSLNISACEKLKCLP 1045
Query: 780 ---LHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFF 836
L L +L L +CP + EG LP NL L I C KL+ R+EW L+ L
Sbjct: 1046 ENMLELLPSLKELRLTNCPEI----EGELPFNLQKLDIRYCKKLLNGRKEWHLQRLTELV 1101
Query: 837 VCDD--FENVESFPKESLLPPTLSYLNLNN--------------CSKLRIMNN------- 873
+ D E++E + LP +++ L ++N LRI+ N
Sbjct: 1102 IHHDGSDEDIEHWE----LPCSITRLEVSNLITLSSQHLKSLTSLQFLRIVGNLSQIQSQ 1157
Query: 874 ---EGFLHLKSLE--------------------FLYIINCPSLERLPEEALPNSLYSLWI 910
F HL SL+ L I NCP+L+ L E ALP+SL L I
Sbjct: 1158 GQLSSFSHLTSLQTLRIRNLQSLAESALPSSLSHLNIYNCPNLQSLSESALPSSLSHLTI 1217
Query: 911 KDCP 914
+CP
Sbjct: 1218 YNCP 1221
Score = 107 bits (268), Expect = 1e-21
Identities = 107/360 (29%), Positives = 159/360 (43%), Gaps = 50/360 (13%)
Query: 596 FLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCL---------- 645
F++ L +S+CD + + EF ++ + V +F + E LC+
Sbjct: 963 FVEYLGVSNCDCVDDMSPEFIPTARKLSIESCHNVTRF-LIPTATETLCIFNCENVEKLS 1021
Query: 646 ---EGFPLLKELYIRECPKLKMSLPQH----LPSLQKLFINDCKMLEASIPNGDNIIDLD 698
G L L I C KLK LP++ LPSL++L + +C +E +P N+ LD
Sbjct: 1022 VACGGAAQLTSLNISACEKLKC-LPENMLELLPSLKELRLTNCPEIEGELPF--NLQKLD 1078
Query: 699 IKRCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPTLDLC 758
I+ C ++L L++L L + + S E I + L+++ + + L
Sbjct: 1079 IRYCKKLLNGRKEWHLQRLTELVIHH-DGSDEDIEHWELPCSITRLEVSNLITLSSQHLK 1137
Query: 759 CYNSLGELSITRWCSSSLSFS-LHLFTNLYSLWFVDCPNLDSFPEGGLPCNLLSLTITNC 817
SL L I S S L F++L SL + NL S E LP +L L I NC
Sbjct: 1138 SLTSLQFLRIVGNLSQIQSQGQLSSFSHLTSLQTLRIRNLQSLAESALPSSLSHLNIYNC 1197
Query: 818 PKLIASRQEWGLKSLKYFFVCDDFENVESFPKESLLPPTLSYLNLNNCSKLRIMNNEGFL 877
P L +SL ES LP +LS+L + NC L+ ++
Sbjct: 1198 PNL---------QSLS----------------ESALPSSLSHLTIYNCPNLQSLSESALP 1232
Query: 878 HLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKYQKEGGEQRDTICHIPCV 937
SL L I NCP+L+ L E ALP+SL LWI CPL++ + GE I HIP +
Sbjct: 1233 --SSLSHLTIYNCPNLQSLSESALPSSLSKLWIFKCPLLRSLLEFVKGEYWPQIAHIPTI 1290
Score = 38.9 bits (89), Expect = 0.74
Identities = 96/437 (21%), Positives = 161/437 (35%), Gaps = 115/437 (26%)
Query: 539 VLEALQPN-RNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTLPFL 597
VL + P R+L+ L++S YK PN + L L L+L + L + L L
Sbjct: 575 VLHNILPTLRSLRVLSLSHYKNKELPNDLF-IKLKLLRFLDLSCTWITKLPDSICGLYNL 633
Query: 598 KMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCLEGFPLLKELYIR 657
+ L +S C ++ + L+ EK+ N L+ L +
Sbjct: 634 ETLLLSSCYKLEELP------------------LQMEKLIN------------LRHLDVS 663
Query: 658 ECPKLKMSLPQHLPSLQKL----------------FINDCKML--EASIPNGDNIID--- 696
+LKM P HL L+ L ++ + + L S+ +N+++
Sbjct: 664 NTRRLKM--PLHLSRLKSLQVLVGAEFLVVGWRMEYLGEAQNLYGSLSVVKLENVVNRRE 721
Query: 697 -LDIKRCDRILVNELPTSLKKLFILENRYTEFSV-EQIFVNSTILEVLELDLNGSLKCPT 754
+ K ++ V +L K I +N TE + +++ + I EV+ G+
Sbjct: 722 AVKAKMREKNHVEQLSLEWSKSSIADNSQTERDILDELHPHKNIKEVVISGYRGT----- 776
Query: 755 LDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPE-GGLPCNLLSLT 813
+ W + L F L L C + S P G LPC L L+
Sbjct: 777 ------------NFPNWVADPL------FVKLVKLSLSYCKDCYSLPALGQLPC-LKFLS 817
Query: 814 ITNCPKL-IASRQEWGLKSLKYFFVCDD---FENVESFPKESLLP----PTLSYLNLNNC 865
+ + + + + +G S K F C + FE++ + + L PTL L++ NC
Sbjct: 818 VKGMHGIRVVTEEFYGRLSSKKPFNCLEKLKFEDMTEWKQWHALGIGEFPTLEKLSIKNC 877
Query: 866 SKLRIMNNEGFLHLKSLEF-------------------------LYIINCPSLERLPEEA 900
+L + F LK LE L I +C S+ P
Sbjct: 878 PELSLERPIQFSSLKRLEVVGCPVVFDDAQLFRFQLEAMKQIEALNISDCNSVTSFPFSI 937
Query: 901 LPNSLYSLWIKDCPLIK 917
LP +L + I CP +K
Sbjct: 938 LPTTLKRIQISGCPKLK 954
>UniRef100_Q9LRR4 Putative disease resistance RPP13-like protein 1 [Arabidopsis
thaliana]
Length = 1054
Score = 464 bits (1195), Expect = e-129
Identities = 304/848 (35%), Positives = 453/848 (52%), Gaps = 88/848 (10%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSS-ADGEDLNLLQHQLQYMLMGK- 58
YND ++ +F K W +VSE FDV +TK + +S S + DL++LQ +L+ L G
Sbjct: 217 YNDQHVRSYFGTKVWAHVSEEFDVFKITKKVYESVTSRPCEFTDLDVLQVKLKERLTGTG 276
Query: 59 -KYLLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLD 117
+LLVLDD+WN + W+LL PF H + GS+I+VTTR + VA +++ + + +LQ L
Sbjct: 277 LPFLLVLDDLWNENFADWDLLRQPFIHAAQGSQILVTTRSQRVA-SIMCAVHVHNLQPLS 335
Query: 118 KSNCWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWIN 177
+CWSLF+ F + + + +IV KC GLPLA+K+LG +LR EW
Sbjct: 336 DGDCWSLFMKTVFGNQEPCLNREIGDLAERIVHKCRGLPLAVKTLGGVLRFEGKVIEWER 395
Query: 178 ILETDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEG 237
+L + +W L N+ VLR+SY+ LP++LKRCF+YCSIFPKGH F+KD++++LWMAEG
Sbjct: 396 VLSSRIWDLPADKSNLLPVLRVSYYYLPAHLKRCFAYCSIFPKGHAFEKDKVVLLWMAEG 455
Query: 238 LLKCCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFS 297
L+ S+++ EE GNE F++L S S Q+ T Y+MHD +N+L + SGEFS
Sbjct: 456 FLQQTRSSKNLEELGNEYFSELESRSLLQK-------TKTRYIMHDFINELAQFASGEFS 508
Query: 298 IQIEDARVERSVERTRHIWFSLQSNSVDKL-------LELTCEGLHSLILEGTRAMLISN 350
+ ED + ERTR++ + L+ N + + ++ L + +R+ +
Sbjct: 509 SKFEDGCKLQVSERTRYLSY-LRDNYAEPMEFEALREVKFLRTFLPLSLTNSSRSCCLDQ 567
Query: 351 NVQQDLFSRLNFLRMLSFRGCGLLEL-VDEISNLKLLRYLDLSYTWIEILPDTICMLHNL 409
V + L L LR+LS + L D N+ R+LDLS T +E LP ++C ++NL
Sbjct: 568 MVSEKLLPTLTRLRVLSLSHYKIARLPPDFFKNISHARFLDLSRTELEKLPKSLCYMYNL 627
Query: 410 QTLLLEGCCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFIVE 469
QTLLL C L ELP++ S L+NLR+L L ++ MP+ G+L +LQ+L+ F V
Sbjct: 628 QTLLLSYCSSLKELPTDISNLINLRYLDLIGTK----LRQMPRRFGRLKSLQTLTTFFVS 683
Query: 470 EQNVSDLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMD 529
+ S + EL L+ LHG + I L V D+AD+A NL K+L E+ + G +
Sbjct: 684 ASDGSRISELGGLHDLHGKLKIVELQRVVDVADAAEANLNSKKHLREIDFVWRTGSSSSE 743
Query: 530 ES----MAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLC 585
+ ++ V E L+P+R++++L I +YKG FP+W+ +V + L+ C C
Sbjct: 744 NNTNPHRTQNEAEVFEKLRPHRHIEKLAIERYKGRRFPDWLSDPSFSRIVCIRLRECQYC 803
Query: 586 SLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDS-----SSINVLFRSLEVLKFEKMNNWE 640
+ LP LG LP LK L IS G++ IG +FY S FRSLE L+F+ + +W+
Sbjct: 804 TSLPSLGQLPCLKELHISGMVGLQSIGRKFYFSDQQLRDQDQQPFRSLETLRFDNLPDWQ 863
Query: 641 EWLCL-----EGFPLLKELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIPNGD--- 692
EWL + + FP LK+L+I CP+L +LP LPSL L I C +L+ + +
Sbjct: 864 EWLDVRVTRGDLFPSLKKLFILRCPELTGTLPTFLPSLISLHIYKCGLLDFQPDHHEYSY 923
Query: 693 -NIIDLDIK-RCDRILVNELPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSL 750
N+ L IK CD ++ +F + F N LEV + SL
Sbjct: 924 RNLQTLSIKSSCDTLV-------------------KFPLNH-FANLDKLEVDQCTSLYSL 963
Query: 751 KCPTLDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYSLWFVDCPNLDSFPE-GGLPCNL 809
+ L N+L L I DC NL P+ LP N
Sbjct: 964 ELSNEHLRGPNALRNLRIN-----------------------DCQNLQLLPKLNALPQN- 999
Query: 810 LSLTITNC 817
L +TITNC
Sbjct: 1000 LQVTITNC 1007
>UniRef100_Q6F2C2 Putative disease resistance protein [Solanum demissum]
Length = 1312
Score = 461 bits (1187), Expect = e-128
Identities = 323/984 (32%), Positives = 500/984 (49%), Gaps = 122/984 (12%)
Query: 1 YNDNKIKEHFELKAWVYVSESFDVVGLTKAILKSFNSSADGEDLNLLQHQLQYMLMGKKY 60
YND K+K HF LKAW+ VSE +D++ +TK +L+ F D +LN LQ +L+ L GKK+
Sbjct: 222 YNDEKVKNHFGLKAWICVSEPYDILRITKELLQEFGLMVDN-NLNQLQVKLKESLKGKKF 280
Query: 61 LLVLDDIWNGDAERWELLLLPFNHGSFGSKIVVTTREKEVADNVLKSTELFDLQQLDKSN 120
L+VLDD+WN + + W+ L F G GSKI+VTTR++ VA ++ ++ L
Sbjct: 281 LIVLDDVWNENYKEWDDLRNLFVQGDVGSKIIVTTRKESVA--LMMGCGAINVGTLSSEV 338
Query: 121 CWSLFVTHAFQGKSVSEYPNLESVGRKIVEKCGGLPLAIKSLGQLLRKTFSEHEWINILE 180
W LF H+F+ + ++P LE VG +I KC GLPLA+K+L +LR EW +IL
Sbjct: 339 SWDLFKRHSFENRDPEDHPELEEVGIQIAHKCKGLPLALKALAGILRSKSEVDEWRDILR 398
Query: 181 TDMWRLSKVDHNVNSVLRLSYHNLPSNLKRCFSYCSIFPKGHKFKKDELIMLWMAEGLLK 240
+++W L + + L LSY++L LKRCF++C+I+PK + F K+++I LW+A GL++
Sbjct: 399 SEIWELQSCSNGILPALMLSYNDLHPQLKRCFAFCAIYPKDYLFCKEQVIHLWIANGLVQ 458
Query: 241 CCGSNRSEEEFGNESFADLVSISFFQQSFDEIYDTYEHYVMHDLVNDLTKSVSGEFSIQI 300
S N F +L S S F++ + ++MHDLVNDL + S I++
Sbjct: 459 QLHS-------ANHYFLELRSRSLFEKVQESSEWNPGEFLMHDLVNDLAQIASSNLCIRL 511
Query: 301 EDARVERSVERTRHIWFSLQSNSVDKLLEL-TCEGLHSLILEGTR--AMLISNNVQQDLF 357
E+ +E++RHI +S+ + KL L E L +L+ + + +S + D+
Sbjct: 512 EENLGSHMLEQSRHISYSMGLDDFKKLKPLYKLEQLRTLLPINIQQHSYCLSKRILHDIL 571
Query: 358 SRLNFLRMLSFRGCGLLELVDEI-SNLKLLRYLDLSYTWIEILPDTICMLHNLQTLLLEG 416
RL LR LS + EL +++ LK LR+LD S+T I+ LPD+IC+L+NL+TLLL
Sbjct: 572 PRLTSLRALSLSHYSIEELPNDLFIKLKYLRFLDFSWTKIKKLPDSICLLYNLETLLLSH 631
Query: 417 CCELTELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSL--SYFIVEEQNVS 474
C L ELP + KL+NLRHL + T P H KL +L +L + I+ +
Sbjct: 632 CSYLKELPLHMEKLINLRHLDISE-----AYLTTPLHLSKLKSLHALVGANLILSGRGGL 686
Query: 475 DLKELAKLNHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGREEMDESMAE 534
+++L ++++L+G++ I L NV D +S N+++ K++E L +++ G + ++ E
Sbjct: 687 RMEDLGEVHNLYGSLSILELQNVVDRRESLKANMREKKHVERLSLEWSGSNADNSQTERE 746
Query: 535 SNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCGLCSLLPPLGTL 594
+L+ LQPN N+K L+ ++L +C C LP LG L
Sbjct: 747 ----ILDELQPNTNIK----------------------ELIEMSLSYCKDCDSLPALGQL 780
Query: 595 PFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLCL--EGFPLLK 652
P LK L+I I + EEFY S S F+SLE L+F M W++W L FP+L+
Sbjct: 781 PCLKFLTIRGMHQITEVSEEFYGSLSSTKPFKSLEKLEFAGMPEWKQWHVLGKGEFPILE 840
Query: 653 ELYIRECPKLKMSLPQHLPSLQKLFINDCKMLEASIP-NGDNIIDLDIKRCDRI------ 705
EL+I CPKL LP++L SL++L I++C L P N+ +L + C ++
Sbjct: 841 ELWINGCPKLIGKLPENLSSLRRLRISECPELSLETPIQLSNLKELKVADCPKVGVLFAN 900
Query: 706 -------------LVNELPTSLKKL-----FILENRYTEFSVE---QIFVNSTILEVLEL 744
+V + T K L L++R E + +N+ LE +L
Sbjct: 901 AQLFTSQLEGMKQIVKLVITDCKSLTSLPISTLKSREISGCGELKLEASMNAMFLE--DL 958
Query: 745 DLNGS------LKCPTLDLCCYNSLGELSITRW--------CSSSLSFSLHLFTNLYSLW 790
L G + L + N+L L I C + S+ + SL
Sbjct: 959 SLKGCDSPELFPRARNLSVRSCNNLTRLLIPTETETLSFGDCDNLEILSVACGIQMTSLN 1018
Query: 791 FVDCPNLDSFPE-------------------------GGLPCNLLSLTITNCPKLIASRQ 825
+C L S PE GGLP NL L I+ C KL+ R+
Sbjct: 1019 IHNCQKLKSLPEHMQELLPSLKELTLDNCPEIESFPQGGLPFNLQFLWISRCKKLVNGRK 1078
Query: 826 EWGLKSLKYFFVCDDFENVESFPKESL-LPPTLSYLNLNNCSKLRIMNNEGFLHLKSLEF 884
EW L+ L + + E+ LP ++ L + N L+ ++++ L SLE+
Sbjct: 1079 EWHLQRLPSLMQLEISHDGSDIAGENWELPCSIRRLTIAN---LKTLSSQLLKSLTSLEY 1135
Query: 885 LYIINCPSLERLPEEALPNSLYSL 908
LY IN P ++ L EE LP+SL L
Sbjct: 1136 LYAINLPQIQSLLEEELPSSLSEL 1159
Score = 117 bits (294), Expect = 1e-24
Identities = 157/557 (28%), Positives = 238/557 (42%), Gaps = 118/557 (21%)
Query: 409 LQTLLLEGCCELT-ELPSNFSKLVNLRHLKLPSHNGRPCIKTMPKHTGKLNNLQSLSYFI 467
L+ L + GC +L +LP N S L LR + P + I+ L+NL+ L
Sbjct: 839 LEELWINGCPKLIGKLPENLSSLRRLRISECPELSLETPIQ--------LSNLKELK--- 887
Query: 468 VEEQNVSDLKELAKL--NHLHGAIDIEGLGNVSDLADSATVNLKDTKYLEELHMKFDGGR 525
V+D ++ L N +EG+ + L + D K L L + R
Sbjct: 888 -----VADCPKVGVLFANAQLFTSQLEGMKQIVKLV------ITDCKSLTSLPISTLKSR 936
Query: 526 EEMDESMAESNVSVLEALQPNRNLKRLTISKYKGNSFPNWIRGYHLPNLVSLNLQFCG-L 584
E S L+ L+ + N L KG P P +L+++ C L
Sbjct: 937 EI-------SGCGELK-LEASMNAMFLEDLSLKGCDSPELF-----PRARNLSVRSCNNL 983
Query: 585 CSLLPPLGTLPFLKMLSISDCDGIKIIGEEFYDSSSINVLFRSLEVLKFEKMNNWEEWLC 644
LL P T + LS DCD ++I+ S + + SL + +K+ + E +
Sbjct: 984 TRLLIPTET----ETLSFGDCDNLEIL------SVACGIQMTSLNIHNCQKLKSLPEHM- 1032
Query: 645 LEGFPLLKELYIRECPKLKMSLPQH-LP-SLQKLFINDCKML-----EASIPNGDNIIDL 697
E P LKEL + CP+++ S PQ LP +LQ L+I+ CK L E + +++ L
Sbjct: 1033 QELLPSLKELTLDNCPEIE-SFPQGGLPFNLQFLWISRCKKLVNGRKEWHLQRLPSLMQL 1091
Query: 698 DIKRCDRILVNE---LPTSLKKLFILENRYTEFSVEQIFVNSTILEVLELDLNGSLKCPT 754
+I + E LP S+++L I + Q+ + T LE L
Sbjct: 1092 EISHDGSDIAGENWELPCSIRRLTIANLKTLS---SQLLKSLTSLEYLY----------A 1138
Query: 755 LDLCCYNSLGELSITRWCSSSLSFSLHLFTNLYSL---------WFV-----DCPNLDSF 800
++L SL E + SS LH +L+SL WF DCPNL S
Sbjct: 1139 INLPQIQSLLEEELP---SSLSELHLHQHHDLHSLPTEGLQRLMWFRCLEIWDCPNLQSL 1195
Query: 801 PEGGLPCNLLSLTITNCPKLIASRQEWGLKSLKYFFVCDDFENVESFPKESLLPPTLSYL 860
PE G+P +L LTI +C N++S P ES +P +LS L
Sbjct: 1196 PESGMPSSLSKLTIQHC------------------------SNLQSLP-ESGMPSSLSDL 1230
Query: 861 NLNNCSKLRIMNNEGFLHLKSLEFLYIINCPSLERLPEEALPNSLYSLWIKDCPLIKVKY 920
++NC L+ + GF SL L I NC +L+ LPE +P S+ +L+I +CPL+K
Sbjct: 1231 TISNCPSLQSLPESGFP--SSLSELGIWNCSNLQSLPESGMPPSICNLYISECPLLKPLL 1288
Query: 921 QKEGGEQRDTICHIPCV 937
+ G+ I HIP +
Sbjct: 1289 EFNKGDYWPKIAHIPTI 1305
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.321 0.139 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,616,550,892
Number of Sequences: 2790947
Number of extensions: 70145577
Number of successful extensions: 184157
Number of sequences better than 10.0: 2946
Number of HSP's better than 10.0 without gapping: 1209
Number of HSP's successfully gapped in prelim test: 1772
Number of HSP's that attempted gapping in prelim test: 169371
Number of HSP's gapped (non-prelim): 7769
length of query: 946
length of database: 848,049,833
effective HSP length: 137
effective length of query: 809
effective length of database: 465,690,094
effective search space: 376743286046
effective search space used: 376743286046
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 80 (35.4 bits)
Medicago: description of AC146704.8


