

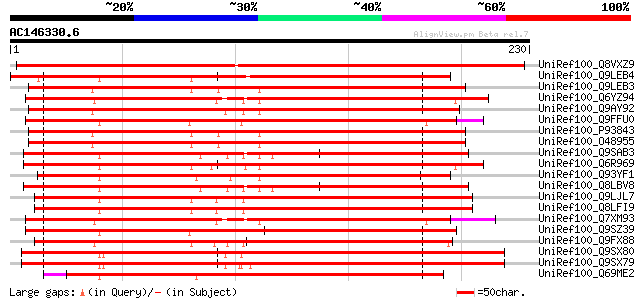
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146330.6 + phase: 0
(230 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q8VXZ9 Putative DNA binding protein ACBF [Arabidopsis ... 355 7e-97
UniRef100_Q9LEB4 RNA Binding Protein 45 [Nicotiana plumbaginifolia] 278 1e-73
UniRef100_Q9LEB3 RNA Binding Protein 47 [Nicotiana plumbaginifolia] 268 6e-71
UniRef100_Q6YZ94 Putative RNA Binding Protein 45 [Oryza sativa] 268 1e-70
UniRef100_Q9AY92 Putative RNA binding protein [Oryza sativa] 267 1e-70
UniRef100_Q9FFU0 Similarity to polyadenylate-binding protein 5 [... 267 2e-70
UniRef100_P93843 DNA binding protein ACBF [Nicotiana tabacum] 266 2e-70
UniRef100_O48955 Putative RNA binding protein [Nicotiana tabacum] 266 2e-70
UniRef100_Q9SAB3 F25C20.21 protein [Arabidopsis thaliana] 262 6e-69
UniRef100_Q6R969 DNA-binding protein [Lycopersicon esculentum] 260 2e-68
UniRef100_Q93YF1 Nucleic acid binding protein [Nicotiana tabacum] 257 1e-67
UniRef100_Q8LBV8 Putative DNA binding protein [Arabidopsis thali... 257 1e-67
UniRef100_Q9LJL7 DNA/RNA binding protein-like [Arabidopsis thali... 256 2e-67
UniRef100_Q8LFI9 Nuclear acid binding protein, putative [Arabido... 256 2e-67
UniRef100_Q7XM93 OSJNBb0060E08.6 protein [Oryza sativa] 256 2e-67
UniRef100_Q9SZ39 Putative DNA binding protein [Arabidopsis thali... 255 7e-67
UniRef100_Q9FX88 Putative RNA binding protein [Arabidopsis thali... 251 1e-65
UniRef100_Q9SX80 F16N3.23 protein [Arabidopsis thaliana] 244 1e-63
UniRef100_Q9SX79 F16N3.24 protein [Arabidopsis thaliana] 242 6e-63
UniRef100_Q69ME2 Putative nucleic acid binding protein [Oryza sa... 239 4e-62
>UniRef100_Q8VXZ9 Putative DNA binding protein ACBF [Arabidopsis thaliana]
Length = 425
Score = 355 bits (910), Expect = 7e-97
Identities = 169/225 (75%), Positives = 188/225 (83%), Gaps = 1/225 (0%)
Query: 4 TYHQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEF 63
+YH P +LEEVRTLWIGDLQYWVDENYLT CFS TGE++S+K+IRNKITGQPEGYGFIEF
Sbjct: 13 SYHHPQTLEEVRTLWIGDLQYWVDENYLTSCFSQTGELVSVKVIRNKITGQPEGYGFIEF 72
Query: 64 VSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYL 123
+SH+AAER LQTYNGTQMPGTE TFRLNWASFG G++ DAGPDHSIFVGDLAPDVTDYL
Sbjct: 73 ISHAAAERTLQTYNGTQMPGTELTFRLNWASFGSGQK-VDAGPDHSIFVGDLAPDVTDYL 131
Query: 124 LQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRIS 183
LQETFR HY SVRGAKVVTDP+TGRSKGYGFVKF++ESERNRAM+EMNG+YCSTRPMRIS
Sbjct: 132 LQETFRVHYSSVRGAKVVTDPSTGRSKGYGFVKFAEESERNRAMAEMNGLYCSTRPMRIS 191
Query: 184 AATPKKTTGYQQNPYAAVVAAAPVPKGITKPYTFFIIRIISIIKC 228
AATPKK G QQ V VP + P ++ S + C
Sbjct: 192 AATPKKNVGVQQQYVTKAVYPVTVPSAVAAPVQAYVAPPESDVTC 236
Score = 45.1 bits (105), Expect = 0.001
Identities = 47/187 (25%), Positives = 77/187 (41%), Gaps = 33/187 (17%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
P S T+ + +L V E L FS GEVI +KI K GYG+++F +
Sbjct: 230 PESDVTCTTISVANLDQNVTEEELKKAFSQLGEVIYVKIPATK------GYGYVQFKTRP 283
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQET 127
+AE +Q G Q+ G +Q R++W+ + G D + D
Sbjct: 284 SAEEAVQRMQG-QVIG-QQAVRISWSK--------NPGQDGWVTQAD----------PNQ 323
Query: 128 FRTHYGSVRGAK-----VVTDPNTGRSKGYGFVKFSDESERNRAMSE--MNGVYCSTRPM 180
+ +YG +G DP+ GYG+ ++ + E + +S GV + + +
Sbjct: 324 WNGYYGYGQGYDAYAYGATQDPSVYAYGGYGYPQYPQQGEGTQDISNSAAGGVAGAEQEL 383
Query: 181 RISAATP 187
ATP
Sbjct: 384 YDPLATP 390
>UniRef100_Q9LEB4 RNA Binding Protein 45 [Nicotiana plumbaginifolia]
Length = 409
Score = 278 bits (710), Expect = 1e-73
Identities = 138/198 (69%), Positives = 158/198 (79%), Gaps = 4/198 (2%)
Query: 1 MATTYHQPASL---EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEG 57
MATT P+ EVR+LWIGDLQYW+DENYL+ CF HTGE++S K+IRNK TGQ EG
Sbjct: 67 MATTNPNPSPTGNPNEVRSLWIGDLQYWMDENYLSTCFYHTGELVSAKVIRNKQTGQSEG 126
Query: 58 YGFIEFVSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAP 117
YGF+EF SH+AAE +LQTYNGT MP EQ FR+NWAS G GERR D+ +H+IFVGDLA
Sbjct: 127 YGFLEFRSHAAAETILQTYNGTLMPNVEQNFRMNWASLGAGERRDDSA-EHTIFVGDLAA 185
Query: 118 DVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCST 177
DVTDY+LQETF++ Y SVRGAKVVTD TGRSKGYGFVKF+DESE+ RAM+EMNGV CST
Sbjct: 186 DVTDYILQETFKSVYSSVRGAKVVTDRITGRSKGYGFVKFADESEQLRAMTEMNGVLCST 245
Query: 178 RPMRISAATPKKTTGYQQ 195
RPMRI A KK G Q
Sbjct: 246 RPMRIGPAANKKPVGTPQ 263
Score = 64.3 bits (155), Expect = 2e-09
Identities = 51/184 (27%), Positives = 82/184 (43%), Gaps = 26/184 (14%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F V K++ ++ITG+ +GYGF++F S R +
Sbjct: 177 TIFVGDLAADVTDYILQETFKSVYSSVRGAKVVTDRITGRSKGYGFVKFADESEQLRAMT 236
Query: 75 TYNGT---------------QMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDV 119
NG + GT Q GE P+ + +IFVG L P V
Sbjct: 237 EMNGVLCSTRPMRIGPAANKKPVGTPQKATYQNPQATQGESDPN---NTTIFVGGLDPTV 293
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
+ L++ F + YG + K+V K GFV+F + +A+S +NG +
Sbjct: 294 AEEHLRQVF-SPYGELVHVKIVA------GKRCGFVQFGTRASAEQALSSLNGTQLGGQS 346
Query: 180 MRIS 183
+R+S
Sbjct: 347 IRLS 350
Score = 52.0 bits (123), Expect = 1e-05
Identities = 28/77 (36%), Positives = 46/77 (59%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V E +L FS GE++ +KI+ K GF++F + ++AE+ L +
Sbjct: 283 TIFVGGLDPTVAEEHLRQVFSPYGELVHVKIVAGK------RCGFVQFGTRASAEQALSS 336
Query: 76 YNGTQMPGTEQTFRLNW 92
NGTQ+ G Q+ RL+W
Sbjct: 337 LNGTQLGG--QSIRLSW 351
>UniRef100_Q9LEB3 RNA Binding Protein 47 [Nicotiana plumbaginifolia]
Length = 428
Score = 268 bits (686), Expect = 6e-71
Identities = 127/194 (65%), Positives = 155/194 (79%)
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
+S E+ +T+WIGDLQ W+DE+YL CFS GEVIS+KIIRNK TGQ E YGF+EF +H+A
Sbjct: 78 SSSEDNKTIWIGDLQQWMDESYLHSCFSQAGEVISVKIIRNKQTGQSERYGFVEFNTHAA 137
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETF 128
AE+VLQ+YNGT MP TEQ FRLNWA F GE+R + G D SIFVGDLA DVTD +L++TF
Sbjct: 138 AEKVLQSYNGTMMPNTEQPFRLNWAGFSTGEKRAETGSDFSIFVGDLASDVTDTMLRDTF 197
Query: 129 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
+ Y S++GAKVV D NTG SKGYGFV+F DESER+RAM+EMNGVYCS+R MRI ATPK
Sbjct: 198 ASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMTEMNGVYCSSRAMRIGVATPK 257
Query: 189 KTTGYQQNPYAAVV 202
K + ++Q AV+
Sbjct: 258 KPSAHEQYSSQAVI 271
Score = 60.5 bits (145), Expect = 3e-08
Identities = 51/193 (26%), Positives = 82/193 (42%), Gaps = 32/193 (16%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F S + K++ + TG +GYGF+ F S R +
Sbjct: 178 SIFVGDLASDVTDTMLRDTFASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMT 237
Query: 75 TYNGT--------------QMPGTEQTFRLN-------WASFGIGERRPDAGPDHS---I 110
NG + P + + +AS G + D S I
Sbjct: 238 EMNGVYCSSRAMRIGVATPKKPSAHEQYSSQAVILSGGYASNGAATHGSQSDGDSSNTTI 297
Query: 111 FVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 170
FVG L +VTD L+++F +G V K+ KG GFV+FSD S A+ ++
Sbjct: 298 FVGGLDSEVTDEELRQSFN-QFGEVVSVKIPA------GKGCGFVQFSDRSSAQEAIQKL 350
Query: 171 NGVYCSTRPMRIS 183
+G + +R+S
Sbjct: 351 SGAIIGKQAVRLS 363
>UniRef100_Q6YZ94 Putative RNA Binding Protein 45 [Oryza sativa]
Length = 427
Score = 268 bits (684), Expect = 1e-70
Identities = 136/206 (66%), Positives = 158/206 (76%), Gaps = 4/206 (1%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
PA EVRTLWIGDLQYW+DENY++ CF+ TGE+ S+K+IR+K TGQ +GYGFIEF SH+
Sbjct: 88 PAGPNEVRTLWIGDLQYWMDENYISACFAPTGELQSVKLIRDKQTGQLQGYGFIEFTSHA 147
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQET 127
AERVLQTYNG MP EQT+RLNWAS GE+R D PD++IFVGDLA DVTDY+LQET
Sbjct: 148 GAERVLQTYNGAMMPNVEQTYRLNWAS--AGEKRDDT-PDYTIFVGDLAADVTDYILQET 204
Query: 128 FRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATP 187
FR HY SV+GAKVVTD T RSKGYGFVKF D SE+ RAM+EMNG+ CS+RPMRI A
Sbjct: 205 FRVHYPSVKGAKVVTDKMTMRSKGYGFVKFGDPSEQARAMTEMNGMVCSSRPMRIGPAAN 264
Query: 188 KKTTGYQQN-PYAAVVAAAPVPKGIT 212
KK TG Q+ P A V + P T
Sbjct: 265 KKATGVQEKVPSAQGVQSDSDPSNTT 290
Score = 67.0 bits (162), Expect = 4e-10
Identities = 54/181 (29%), Positives = 90/181 (48%), Gaps = 22/181 (12%)
Query: 16 TLWIGDLQYWVDENYLTHCFS-HTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F H V K++ +K+T + +GYGF++F S R +
Sbjct: 186 TIFVGDLAADVTDYILQETFRVHYPSVKGAKVVTDKMTMRSKGYGFVKFGDPSEQARAMT 245
Query: 75 TYNGTQMPGTEQTFRL----NWASFGIGERRP-------DAGPDHS-IFVGDLAPDVTDY 122
NG M + + R+ N + G+ E+ P D+ P ++ IFVG L P VTD
Sbjct: 246 EMNG--MVCSSRPMRIGPAANKKATGVQEKVPSAQGVQSDSDPSNTTIFVGGLDPSVTDD 303
Query: 123 LLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRI 182
+L++ F T YG V K+ K GFV+F++ + + A+ + G + +R+
Sbjct: 304 MLKQVF-TPYGDVVHVKIPV------GKRCGFVQFANRASADEALVLLQGTLIGGQNVRL 356
Query: 183 S 183
S
Sbjct: 357 S 357
>UniRef100_Q9AY92 Putative RNA binding protein [Oryza sativa]
Length = 402
Score = 267 bits (683), Expect = 1e-70
Identities = 129/191 (67%), Positives = 152/191 (79%)
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
A+ +EVRTLWIGDLQ+W++ENYL +CFS GE+IS KIIRNK TGQPEGYGFIEF SH+
Sbjct: 61 AAGDEVRTLWIGDLQFWMEENYLYNCFSQAGELISAKIIRNKQTGQPEGYGFIEFGSHAI 120
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETF 128
AE+VLQ YNG MP Q F+LNWA+ G GE+R D G D++IFVGDLA DVTD +LQ+TF
Sbjct: 121 AEQVLQGYNGQMMPNGNQVFKLNWATSGAGEKRGDDGSDYTIFVGDLASDVTDLILQDTF 180
Query: 129 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
+ HY SV+GAKVV D +TGRSKGYGFVKF D E+ RAM+EMNG YCS+RPMRI A+ K
Sbjct: 181 KAHYQSVKGAKVVFDRSTGRSKGYGFVKFGDLDEQTRAMTEMNGQYCSSRPMRIGPASNK 240
Query: 189 KTTGYQQNPYA 199
K G QQ P A
Sbjct: 241 KNIGGQQQPSA 251
Score = 59.7 bits (143), Expect = 6e-08
Identities = 51/186 (27%), Positives = 91/186 (48%), Gaps = 28/186 (15%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F +H V K++ ++ TG+ +GYGF++F R +
Sbjct: 161 TIFVGDLASDVTDLILQDTFKAHYQSVKGAKVVFDRSTGRSKGYGFVKFGDLDEQTRAMT 220
Query: 75 TYNGTQMPGTEQTFRLNWAS----FGIGERRP------------DAGPDHS-IFVGDLAP 117
NG + + R+ AS G G+++P D+ P+++ +FVG L P
Sbjct: 221 EMNGQYC--SSRPMRIGPASNKKNIG-GQQQPSATYQNTQGTDSDSDPNNTTVFVGGLDP 277
Query: 118 DVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCST 177
VTD +L++ F + YG + K+ K GFV++S+ + A+ +NG
Sbjct: 278 SVTDEVLKQAF-SPYGELVYVKIPV------GKRCGFVQYSNRASAEEAIRMLNGSQLGG 330
Query: 178 RPMRIS 183
+ +R+S
Sbjct: 331 QSIRLS 336
>UniRef100_Q9FFU0 Similarity to polyadenylate-binding protein 5 [Arabidopsis
thaliana]
Length = 390
Score = 267 bits (682), Expect = 2e-70
Identities = 128/192 (66%), Positives = 149/192 (76%), Gaps = 1/192 (0%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
P S +V++LWIGDLQ W+DENY+ F+ +GE S K+IRNK+TGQ EGYGFIEFVSHS
Sbjct: 53 PGSASDVKSLWIGDLQQWMDENYIMSVFAQSGEATSAKVIRNKLTGQSEGYGFIEFVSHS 112
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERR-PDAGPDHSIFVGDLAPDVTDYLLQE 126
AERVLQTYNG MP TEQTFRLNWA G GE+R GPDH+IFVGDLAP+VTDY+L +
Sbjct: 113 VAERVLQTYNGAPMPSTEQTFRLNWAQAGAGEKRFQTEGPDHTIFVGDLAPEVTDYMLSD 172
Query: 127 TFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAAT 186
TF+ YGSV+GAKVV D TGRSKGYGFV+F+DE+E+ RAM+EMNG YCSTRPMRI A
Sbjct: 173 TFKNVYGSVKGAKVVLDRTTGRSKGYGFVRFADENEQMRAMTEMNGQYCSTRPMRIGPAA 232
Query: 187 PKKTTGYQQNPY 198
K Q Y
Sbjct: 233 NKNALPMQPAMY 244
Score = 59.7 bits (143), Expect = 6e-08
Identities = 52/213 (24%), Positives = 93/213 (43%), Gaps = 29/213 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L+ F + G V K++ ++ TG+ +GYGF+ F + R +
Sbjct: 155 TIFVGDLAPEVTDYMLSDTFKNVYGSVKGAKVVLDRTTGRSKGYGFVRFADENEQMRAMT 214
Query: 75 TYNG----------------TQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPD 118
NG +P ++ N G+ P+ + +IFVG L +
Sbjct: 215 EMNGQYCSTRPMRIGPAANKNALPMQPAMYQ-NTQGANAGDNDPN---NTTIFVGGLDAN 270
Query: 119 VTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTR 178
VTD L+ F +G + K+ K GFV++++++ A+S +NG +
Sbjct: 271 VTDDELKSIFG-QFGELLHVKIPP------GKRCGFVQYANKASAEHALSVLNGTQLGGQ 323
Query: 179 PMRIS-AATPKKTTGYQQNPYAAVVAAAPVPKG 210
+R+S +P K + Q P P+G
Sbjct: 324 SIRLSWGRSPNKQSDQAQWNGGGYYGYPPQPQG 356
>UniRef100_P93843 DNA binding protein ACBF [Nicotiana tabacum]
Length = 428
Score = 266 bits (681), Expect = 2e-70
Identities = 127/194 (65%), Positives = 153/194 (78%)
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
+S E+ +T+WIGDLQ W+DE+YL CFS GEVIS+KIIRNK TGQ E YGF+EF +H+A
Sbjct: 78 SSSEDNKTIWIGDLQQWMDESYLHSCFSQAGEVISVKIIRNKQTGQSERYGFVEFNTHAA 137
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETF 128
AE+VLQ+YNGT MP EQ FRLNWA F GE+R + G D SIFVGDLA DVTD +L++TF
Sbjct: 138 AEKVLQSYNGTMMPNAEQPFRLNWAGFSTGEKRAETGSDFSIFVGDLASDVTDTMLRDTF 197
Query: 129 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
+ Y S++GAKVV D NTG SKGYGFV+F DESER+RAM+EMNGVYCS+R MRI ATPK
Sbjct: 198 ASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMTEMNGVYCSSRAMRIGVATPK 257
Query: 189 KTTGYQQNPYAAVV 202
K + QQ AV+
Sbjct: 258 KPSAQQQYSSQAVI 271
Score = 63.5 bits (153), Expect = 4e-09
Identities = 53/193 (27%), Positives = 83/193 (42%), Gaps = 32/193 (16%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F S + K++ + TG +GYGF+ F S R +
Sbjct: 178 SIFVGDLASDVTDTMLRDTFASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMT 237
Query: 75 TYNGT--------------QMPGTEQTFRLN-------WASFGIGERRPDAGPDHS---I 110
NG + P +Q + +AS G + D S I
Sbjct: 238 EMNGVYCSSRAMRIGVATPKKPSAQQQYSSQAVILSGGYASNGAATHGSQSDGDASNTTI 297
Query: 111 FVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 170
FVG L DVTD L+++F +G V K+ KG GFV+FSD S A+ ++
Sbjct: 298 FVGGLDSDVTDEELRQSFN-QFGEVVSVKIPA------GKGCGFVQFSDRSSAQEAIQKL 350
Query: 171 NGVYCSTRPMRIS 183
+G + +R+S
Sbjct: 351 SGAIIGKQAVRLS 363
>UniRef100_O48955 Putative RNA binding protein [Nicotiana tabacum]
Length = 482
Score = 266 bits (681), Expect = 2e-70
Identities = 127/194 (65%), Positives = 153/194 (78%)
Query: 9 ASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSA 68
+S E+ +T+WIGDLQ W+DE+YL CFS GEVIS+KIIRNK TGQ E YGF+EF +H+A
Sbjct: 132 SSSEDNKTIWIGDLQQWMDESYLHSCFSQAGEVISVKIIRNKQTGQSERYGFVEFNTHAA 191
Query: 69 AERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETF 128
AE+VLQ+YNGT MP EQ FRLNWA F GE+R + G D SIFVGDLA DVTD +L++TF
Sbjct: 192 AEKVLQSYNGTMMPNAEQPFRLNWAGFSTGEKRAETGSDFSIFVGDLASDVTDTMLRDTF 251
Query: 129 RTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPK 188
+ Y S++GAKVV D NTG SKGYGFV+F DESER+RAM+EMNGVYCS+R MRI ATPK
Sbjct: 252 ASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMTEMNGVYCSSRAMRIGVATPK 311
Query: 189 KTTGYQQNPYAAVV 202
K + QQ AV+
Sbjct: 312 KPSAQQQYSSQAVI 325
Score = 63.5 bits (153), Expect = 4e-09
Identities = 53/193 (27%), Positives = 83/193 (42%), Gaps = 32/193 (16%)
Query: 16 TLWIGDLQYWVDENYLTHCF-SHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F S + K++ + TG +GYGF+ F S R +
Sbjct: 232 SIFVGDLASDVTDTMLRDTFASRYPSLKGAKVVVDANTGHSKGYGFVRFGDESERSRAMT 291
Query: 75 TYNGT--------------QMPGTEQTFRLN-------WASFGIGERRPDAGPDHS---I 110
NG + P +Q + +AS G + D S I
Sbjct: 292 EMNGVYCSSRAMRIGVATPKKPSAQQQYSSQAVILSGGYASNGAATHGSQSDGDASNTTI 351
Query: 111 FVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 170
FVG L DVTD L+++F +G V K+ KG GFV+FSD S A+ ++
Sbjct: 352 FVGGLDSDVTDEELRQSFN-QFGEVVSVKIPA------GKGCGFVQFSDRSSAQEAIQKL 404
Query: 171 NGVYCSTRPMRIS 183
+G + +R+S
Sbjct: 405 SGAIIGKQAVRLS 417
>UniRef100_Q9SAB3 F25C20.21 protein [Arabidopsis thaliana]
Length = 405
Score = 262 bits (669), Expect = 6e-69
Identities = 129/198 (65%), Positives = 156/198 (78%), Gaps = 2/198 (1%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP + +E+RTLWIGDLQYW+DEN+L CF+HTGE++S K+IRNK TGQ EGYGFIEF SH
Sbjct: 54 QPTTADEIRTLWIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASH 113
Query: 67 SAAERVLQTYNGTQMPG-TEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQ 125
+AAERVLQT+N +P +Q FRLNWAS G++R D+ PD++IFVGDLA DVTDY+L
Sbjct: 114 AAAERVLQTFNNAPIPSFPDQLFRLNWASLSSGDKRDDS-PDYTIFVGDLAADVTDYILL 172
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
ETFR Y SV+GAKVV D TGR+KGYGFV+FSDESE+ RAM+EMNGV CSTRPMRI A
Sbjct: 173 ETFRASYPSVKGAKVVIDRVTGRTKGYGFVRFSDESEQIRAMTEMNGVPCSTRPMRIGPA 232
Query: 186 TPKKTTGYQQNPYAAVVA 203
KK Q++ Y + A
Sbjct: 233 ASKKGVTGQRDSYQSSAA 250
Score = 60.8 bits (146), Expect = 3e-08
Identities = 52/183 (28%), Positives = 88/183 (47%), Gaps = 24/183 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ +++TG+ +GYGF+ F S R +
Sbjct: 156 TIFVGDLAADVTDYILLETFRASYPSVKGAKVVIDRVTGRTKGYGFVRFSDESEQIRAMT 215
Query: 75 TYNGTQMPGTEQTFRLNWASF--GIGERRP-----------DAGPDHS-IFVGDLAPDVT 120
NG +P + + R+ A+ G+ +R D P+++ +FVG L VT
Sbjct: 216 EMNG--VPCSTRPMRIGPAASKKGVTGQRDSYQSSAAGVTTDNDPNNTTVFVGGLDASVT 273
Query: 121 DYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPM 180
D L+ F + YG + K+ K GFV+FS++S A+ +NGV +
Sbjct: 274 DDHLKNVF-SQYGEIVHVKIPA------GKRCGFVQFSEKSCAEEALRMLNGVQLGGTTV 326
Query: 181 RIS 183
R+S
Sbjct: 327 RLS 329
Score = 50.4 bits (119), Expect = 4e-05
Identities = 35/124 (28%), Positives = 56/124 (44%), Gaps = 13/124 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V +++L + FS GE++ +KI K G F++F S AE L+
Sbjct: 262 TVFVGGLDASVTDDHLKNVFSQYGEIVHVKIPAGKRCG------FVQFSEKSCAEEALRM 315
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDL--APDVTDYLLQETFRTHYG 133
NG Q+ GT T RL+W G +G + G + Y + + +YG
Sbjct: 316 LNGVQLGGT--TVRLSW---GRSPSNKQSGDPSQFYYGGYGQGQEQYGYTMPQDPNAYYG 370
Query: 134 SVRG 137
G
Sbjct: 371 GYSG 374
>UniRef100_Q6R969 DNA-binding protein [Lycopersicon esculentum]
Length = 428
Score = 260 bits (665), Expect = 2e-68
Identities = 130/211 (61%), Positives = 157/211 (73%), Gaps = 7/211 (3%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
Q + E+ RT+WIGDLQ W+DE YL CF+ GEVIS+K+IRNK TGQ E YGFIEF +H
Sbjct: 71 QQNASEDNRTIWIGDLQQWMDEGYLHTCFAQAGEVISVKVIRNKQTGQSERYGFIEFNTH 130
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPD------AGPDHSIFVGDLAPDVT 120
AAE+VLQ+YNGT MP EQ FRLNW++F GE+R D +G D SIFVGDLA DVT
Sbjct: 131 EAAEKVLQSYNGTMMPNAEQPFRLNWSAFSSGEKRADVGAGAGSGSDLSIFVGDLASDVT 190
Query: 121 DYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPM 180
D +L++TF + Y SV+GAKVV D NTGRSKGYGFV+F DESER+RAM+EMNG+YCS+R M
Sbjct: 191 DTMLRDTFSSRYPSVKGAKVVIDSNTGRSKGYGFVRFDDESERSRAMTEMNGIYCSSRAM 250
Query: 181 RISAATPKKTTGYQQN-PYAAVVAAAPVPKG 210
RI ATPKK + QQ P A ++A G
Sbjct: 251 RIGVATPKKPSPMQQYFPQAVILAGGHASNG 281
Score = 59.7 bits (143), Expect = 6e-08
Identities = 52/193 (26%), Positives = 84/193 (42%), Gaps = 32/193 (16%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L FS V K++ + TG+ +GYGF+ F S R +
Sbjct: 179 SIFVGDLASDVTDTMLRDTFSSRYPSVKGAKVVIDSNTGRSKGYGFVRFDDESERSRAMT 238
Query: 75 TYNGT--------------QMPGTEQTF-------RLNWASFGIGERRPDAGPDHS---I 110
NG + P Q + AS G + D S +
Sbjct: 239 EMNGIYCSSRAMRIGVATPKKPSPMQQYFPQAVILAGGHASNGAATQTSQTDSDLSNTTV 298
Query: 111 FVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEM 170
FVG L +VTD L+++F + +G+V K+ KG GFV+FS+ S A+ ++
Sbjct: 299 FVGGLDSEVTDEELRQSF-SQFGNVVSVKIPA------GKGCGFVQFSERSAAEDAIEKL 351
Query: 171 NGVYCSTRPMRIS 183
NG + +R+S
Sbjct: 352 NGTVIGAQTVRLS 364
Score = 48.1 bits (113), Expect = 2e-04
Identities = 29/77 (37%), Positives = 41/77 (52%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V + L FS G V+S+KI K G GF++F SAAE ++
Sbjct: 297 TVFVGGLDSEVTDEELRQSFSQFGNVVSVKIPAGK------GCGFVQFSERSAAEDAIEK 350
Query: 76 YNGTQMPGTEQTFRLNW 92
NGT + QT RL+W
Sbjct: 351 LNGTVIGA--QTVRLSW 365
>UniRef100_Q93YF1 Nucleic acid binding protein [Nicotiana tabacum]
Length = 456
Score = 257 bits (657), Expect = 1e-67
Identities = 121/183 (66%), Positives = 150/183 (81%)
Query: 13 EVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERV 72
E RT+W+GDL W+DE+YL +CF+ T EV SIK+IRNK TG EGYGF+EF +H+AAE+V
Sbjct: 115 ENRTIWVGDLLNWMDEDYLRNCFASTNEVASIKVIRNKQTGFSEGYGFVEFFTHAAAEKV 174
Query: 73 LQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHY 132
LQTY+ MP +Q FRLNWA+F +G++R + G D SIFVGDLA DVTD LL ETF T Y
Sbjct: 175 LQTYSCMTMPNVDQPFRLNWATFSMGDKRANNGSDLSIFVGDLAADVTDTLLHETFATKY 234
Query: 133 GSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTG 192
SV+ AKVV D NTGRSKGYGFV+F D++ER++AM+EMNGVYCS+RPMRI AATP+K++G
Sbjct: 235 PSVKAAKVVFDANTGRSKGYGFVRFGDDNERSQAMTEMNGVYCSSRPMRIGAATPRKSSG 294
Query: 193 YQQ 195
YQQ
Sbjct: 295 YQQ 297
Score = 48.1 bits (113), Expect = 2e-04
Identities = 41/187 (21%), Positives = 83/187 (43%), Gaps = 27/187 (14%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F+ V + K++ + TG+ +GYGF+ F + + +
Sbjct: 211 SIFVGDLAADVTDTLLHETFATKYPSVKAAKVVFDANTGRSKGYGFVRFGDDNERSQAMT 270
Query: 75 TYNGTQM-----------PGTEQTFRLNWASFG-------IGERRPDAGPDHS-IFVGDL 115
NG P ++ ++S G +PDA ++ IFVG L
Sbjct: 271 EMNGVYCSSRPMRIGAATPRKSSGYQQQYSSQGGYSNGGPAQGSQPDADSTNTTIFVGGL 330
Query: 116 APDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYC 175
P+V+D L++ F YG + K+ + +F++ ++ A+ ++NG +
Sbjct: 331 DPNVSDEDLRQPF-VQYGEIVSVKIPVEERV------WVWQFANRNDAEEALQKLNGTFI 383
Query: 176 STRPMRI 182
+ +R+
Sbjct: 384 GKQTVRL 390
Score = 34.7 bits (78), Expect = 2.0
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 8/86 (9%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP + T+++G L V + L F GE++S+KI E +F +
Sbjct: 315 QPDADSTNTTIFVGGLDPNVSDEDLRQPFVQYGEIVSVKI------PVEERVWVWQFANR 368
Query: 67 SAAERVLQTYNGTQMPGTEQTFRLNW 92
+ AE LQ NGT + +QT RL W
Sbjct: 369 NDAEEALQKLNGTFI--GKQTVRLFW 392
>UniRef100_Q8LBV8 Putative DNA binding protein [Arabidopsis thaliana]
Length = 404
Score = 257 bits (657), Expect = 1e-67
Identities = 127/198 (64%), Positives = 155/198 (78%), Gaps = 2/198 (1%)
Query: 7 QPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSH 66
QP + +E+RTLWIGDLQYW+DEN+L CF+HTGE++S K+IRNK TGQ EGYGFIEF SH
Sbjct: 53 QPTTADEIRTLWIGDLQYWMDENFLYGCFAHTGEMVSAKVIRNKQTGQVEGYGFIEFASH 112
Query: 67 SAAERVLQTYNGTQMPG-TEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQ 125
+AAERVLQT+N +P +Q FRL WAS G++R D+ PD++IFVGDLA DVTDY+L
Sbjct: 113 AAAERVLQTFNNAPIPSFPDQLFRLXWASLSSGDKRDDS-PDYTIFVGDLAADVTDYILL 171
Query: 126 ETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAA 185
ETFR Y SV+GAKVV + TGR+KGYGFV+FSDESE+ RAM+EMNGV CSTRPMRI A
Sbjct: 172 ETFRASYPSVKGAKVVINRVTGRTKGYGFVRFSDESEQIRAMTEMNGVPCSTRPMRIGPA 231
Query: 186 TPKKTTGYQQNPYAAVVA 203
KK Q++ Y + A
Sbjct: 232 ASKKGVTGQRDSYQSSAA 249
Score = 62.8 bits (151), Expect = 7e-09
Identities = 53/183 (28%), Positives = 88/183 (47%), Gaps = 24/183 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F + V K++ N++TG+ +GYGF+ F S R +
Sbjct: 155 TIFVGDLAADVTDYILLETFRASYPSVKGAKVVINRVTGRTKGYGFVRFSDESEQIRAMT 214
Query: 75 TYNGTQMPGTEQTFRLNWASF--GIGERRP-----------DAGPDHS-IFVGDLAPDVT 120
NG +P + + R+ A+ G+ +R D P+++ +FVG L VT
Sbjct: 215 EMNG--VPCSTRPMRIGPAASKKGVTGQRDSYQSSAAGVTTDNDPNNTTVFVGGLDASVT 272
Query: 121 DYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPM 180
D L+ F + YG + K+ K GFV+FS++S A+ +NGV +
Sbjct: 273 DDHLKNVF-SQYGEIVHVKIPA------GKRCGFVQFSEKSCAEEALRMLNGVQLGGTTV 325
Query: 181 RIS 183
R+S
Sbjct: 326 RLS 328
Score = 50.4 bits (119), Expect = 4e-05
Identities = 35/124 (28%), Positives = 56/124 (44%), Gaps = 13/124 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V +++L + FS GE++ +KI K G F++F S AE L+
Sbjct: 261 TVFVGGLDASVTDDHLKNVFSQYGEIVHVKIPAGKRCG------FVQFSEKSCAEEALRM 314
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDL--APDVTDYLLQETFRTHYG 133
NG Q+ GT T RL+W G +G + G + Y + + +YG
Sbjct: 315 LNGVQLGGT--TVRLSW---GRSPSNKQSGDPSQFYYGGYGQGQEQYGYTMPQDPNAYYG 369
Query: 134 SVRG 137
G
Sbjct: 370 GYSG 373
>UniRef100_Q9LJL7 DNA/RNA binding protein-like [Arabidopsis thaliana]
Length = 489
Score = 256 bits (655), Expect = 2e-67
Identities = 120/195 (61%), Positives = 150/195 (76%), Gaps = 1/195 (0%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
++V+TLW+GDL +W+DE YL CFSHTGEV S+K+IRNK+T Q EGYGF+EF+S +AAE
Sbjct: 105 DDVKTLWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEE 164
Query: 72 VLQTYNGTQMPGTEQTFRLNWASFGIGERRP-DAGPDHSIFVGDLAPDVTDYLLQETFRT 130
VLQ Y+G+ MP ++Q FR+NWASF GE+R + GPD S+FVGDL+PDVTD LL ETF
Sbjct: 165 VLQNYSGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLLHETFSD 224
Query: 131 HYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKT 190
Y SV+ AKVV D NTGRSKGYGFV+F DE+ER+RA++EMNG YCS R MR+ ATPK+
Sbjct: 225 RYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRA 284
Query: 191 TGYQQNPYAAVVAAA 205
QQ + V A
Sbjct: 285 IANQQQHSSQAVILA 299
Score = 61.6 bits (148), Expect = 2e-08
Identities = 52/194 (26%), Positives = 81/194 (40%), Gaps = 33/194 (17%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L FS V S K++ + TG+ +GYGF+ F + R L
Sbjct: 203 SVFVGDLSPDVTDVLLHETFSDRYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALT 262
Query: 75 TYNGT--------------------QMPGTEQTFRL-----NWASFGIGERRPDAGPDHS 109
NG Q + Q L + S G G + + +
Sbjct: 263 EMNGAYCSNRQMRVGIATPKRAIANQQQHSSQAVILAGGHGSNGSMGYGSQSDGESTNAT 322
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
IFVG + PDV D L++ F + +G V K+ KG GFV+F+D A+
Sbjct: 323 IFVGGIDPDVIDEDLRQPF-SQFGEVVSVKIPV------GKGCGFVQFADRKSAEDAIES 375
Query: 170 MNGVYCSTRPMRIS 183
+NG +R+S
Sbjct: 376 LNGTVIGKNTVRLS 389
Score = 43.5 bits (101), Expect = 0.004
Identities = 26/77 (33%), Positives = 42/77 (53%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G + V + L FS GEV+S+KI K G GF++F +AE +++
Sbjct: 322 TIFVGGIDPDVIDEDLRQPFSQFGEVVSVKIPVGK------GCGFVQFADRKSAEDAIES 375
Query: 76 YNGTQMPGTEQTFRLNW 92
NGT + + T RL+W
Sbjct: 376 LNGTVI--GKNTVRLSW 390
>UniRef100_Q8LFI9 Nuclear acid binding protein, putative [Arabidopsis thaliana]
Length = 392
Score = 256 bits (655), Expect = 2e-67
Identities = 120/195 (61%), Positives = 150/195 (76%), Gaps = 1/195 (0%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
++V+TLW+GDL +W+DE YL CFSHTGEV S+K+IRNK+T Q EGYGF+EF+S +AAE
Sbjct: 62 DDVKTLWVGDLLHWMDETYLHSCFSHTGEVSSVKVIRNKLTSQSEGYGFVEFLSRAAAEE 121
Query: 72 VLQTYNGTQMPGTEQTFRLNWASFGIGERRP-DAGPDHSIFVGDLAPDVTDYLLQETFRT 130
VLQ Y+G+ MP ++Q FR+NWASF GE+R + GPD S+FVGDL+PDVTD LL ETF
Sbjct: 122 VLQNYSGSVMPNSDQPFRINWASFSTGEKRAVENGPDLSVFVGDLSPDVTDVLLHETFSD 181
Query: 131 HYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKT 190
Y SV+ AKVV D NTGRSKGYGFV+F DE+ER+RA++EMNG YCS R MR+ ATPK+
Sbjct: 182 RYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALTEMNGAYCSNRQMRVGIATPKRA 241
Query: 191 TGYQQNPYAAVVAAA 205
QQ + V A
Sbjct: 242 IANQQQHSSQAVILA 256
Score = 61.6 bits (148), Expect = 2e-08
Identities = 52/194 (26%), Positives = 81/194 (40%), Gaps = 33/194 (17%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L FS V S K++ + TG+ +GYGF+ F + R L
Sbjct: 160 SVFVGDLSPDVTDVLLHETFSDRYPSVKSAKVVIDSNTGRSKGYGFVRFGDENERSRALT 219
Query: 75 TYNGT--------------------QMPGTEQTFRL-----NWASFGIGERRPDAGPDHS 109
NG Q + Q L + S G G + + +
Sbjct: 220 EMNGAYCSNRQMRVGIATPKRAIANQQQHSSQAVILAGGHGSNGSMGYGSQSDGESTNAT 279
Query: 110 IFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSE 169
IFVG + PDV D L++ F + +G V K+ KG GFV+F+D A+
Sbjct: 280 IFVGGIDPDVIDEDLRQPF-SQFGEVVSVKIPV------GKGCGFVQFADRKSAEDAIES 332
Query: 170 MNGVYCSTRPMRIS 183
+NG +R+S
Sbjct: 333 LNGTVIGKNTVRLS 346
Score = 45.4 bits (106), Expect = 0.001
Identities = 29/90 (32%), Positives = 47/90 (52%), Gaps = 8/90 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G + V + L FS GEV+S+KI K G GF++F +AE +++
Sbjct: 279 TIFVGGIDPDVIDEDLRQPFSQFGEVVSVKIPVGK------GCGFVQFADRKSAEDAIES 332
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAG 105
NGT + + T RL+W + R D+G
Sbjct: 333 LNGTVI--GKNTVRLSWGRSPNKQWRGDSG 360
>UniRef100_Q7XM93 OSJNBb0060E08.6 protein [Oryza sativa]
Length = 414
Score = 256 bits (655), Expect = 2e-67
Identities = 125/188 (66%), Positives = 152/188 (80%), Gaps = 3/188 (1%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
PA+ +EV+TLWIGDLQ W+DE+Y+ +CF+ TGEV S+K+IR+K +GQ +GYGF+EF S +
Sbjct: 72 PAAADEVKTLWIGDLQPWMDESYIYNCFAATGEVQSVKLIRDKQSGQLQGYGFVEFTSRA 131
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQET 127
AA+R+LQTYNG MP E FRLNWAS GE+R D PD++IFVGDLA DVTDYLLQET
Sbjct: 132 AADRILQTYNGQMMPNVEMVFRLNWAS--AGEKRDDT-PDYTIFVGDLAADVTDYLLQET 188
Query: 128 FRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATP 187
FR HY SV+GAKVVTD T RSKGYGFVKF D +E+ RAM+EMNG+ CS+RPMRI A
Sbjct: 189 FRVHYPSVKGAKVVTDKMTMRSKGYGFVKFGDPTEQARAMTEMNGMLCSSRPMRIGPAAN 248
Query: 188 KKTTGYQQ 195
KKTTG Q+
Sbjct: 249 KKTTGVQE 256
Score = 61.6 bits (148), Expect = 2e-08
Identities = 57/215 (26%), Positives = 97/215 (44%), Gaps = 24/215 (11%)
Query: 16 TLWIGDLQYWVDENYLTHCFS-HTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V + L F H V K++ +K+T + +GYGF++F + R +
Sbjct: 170 TIFVGDLAADVTDYLLQETFRVHYPSVKGAKVVTDKMTMRSKGYGFVKFGDPTEQARAMT 229
Query: 75 TYNGTQMPGTEQTFRL----NWASFGIGERRPDAGPDHS--------IFVGDLAPDVTDY 122
NG M + + R+ N + G+ ER P+A S IFVG L P+VT+
Sbjct: 230 EMNG--MLCSSRPMRIGPAANKKTTGVQERVPNAQGAQSENDPNNTTIFVGGLDPNVTED 287
Query: 123 LLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRI 182
+L++ F YG V K+ K GFV++ + +A++ + G + +R+
Sbjct: 288 VLKQVF-APYGEVVHVKIPV------GKRCGFVQYVNRPSAEQALAVLQGTLIGGQNVRL 340
Query: 183 S--AATPKKTTGYQQNPYAAVVAAAPVPKGITKPY 215
S + K + N + A A G + Y
Sbjct: 341 SWGRSLSNKQPQHDSNQWGAGAGAGGYYGGYGQGY 375
>UniRef100_Q9SZ39 Putative DNA binding protein [Arabidopsis thaliana]
Length = 427
Score = 255 bits (651), Expect = 7e-67
Identities = 122/191 (63%), Positives = 143/191 (73%)
Query: 8 PASLEEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHS 67
P S E+R+LWIGDLQ W+DENYL + F TGE + K+IRNK G EGYGFIEFV+H+
Sbjct: 73 PGSAGEIRSLWIGDLQPWMDENYLMNVFGLTGEATAAKVIRNKQNGYSEGYGFIEFVNHA 132
Query: 68 AAERVLQTYNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQET 127
AER LQTYNG MP +EQ FRLNWA G GERR GP+H++FVGDLAPDVTD++L ET
Sbjct: 133 TAERNLQTYNGAPMPSSEQAFRLNWAQLGAGERRQAEGPEHTVFVGDLAPDVTDHMLTET 192
Query: 128 FRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATP 187
F+ Y SV+GAKVV D TGRSKGYGFV+F+DESE+ RAM+EMNG YCS+RPMR A
Sbjct: 193 FKAVYSSVKGAKVVNDRTTGRSKGYGFVRFADESEQIRAMTEMNGQYCSSRPMRTGPAAN 252
Query: 188 KKTTGYQQNPY 198
KK Q Y
Sbjct: 253 KKPLTMQPASY 263
Score = 54.7 bits (130), Expect = 2e-06
Identities = 47/183 (25%), Positives = 80/183 (43%), Gaps = 25/183 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
T+++GDL V ++ LT F V K++ ++ TG+ +GYGF+ F S R +
Sbjct: 174 TVFVGDLAPDVTDHMLTETFKAVYSSVKGAKVVNDRTTGRSKGYGFVRFADESEQIRAMT 233
Query: 75 TYNG--------------TQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVT 120
NG + P T Q GE P + +IFVG + VT
Sbjct: 234 EMNGQYCSSRPMRTGPAANKKPLTMQPASYQNTQGNSGESDPT---NTTIFVGAVDQSVT 290
Query: 121 DYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPM 180
+ L+ F +G + K+ K GFV++++ + +A+S +NG + +
Sbjct: 291 EDDLKSVFG-QFGELVHVKIPA------GKRCGFVQYANRACAEQALSVLNGTQLGGQSI 343
Query: 181 RIS 183
R+S
Sbjct: 344 RLS 346
Score = 45.8 bits (107), Expect = 9e-04
Identities = 29/98 (29%), Positives = 48/98 (48%), Gaps = 11/98 (11%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G + V E+ L F GE++ +KI K G F+++ + + AE+ L
Sbjct: 279 TIFVGAVDQSVTEDDLKSVFGQFGELVHVKIPAGKRCG------FVQYANRACAEQALSV 332
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAGPDHSIFVG 113
NGTQ+ G Q+ RL+W G PD + + G
Sbjct: 333 LNGTQLGG--QSIRLSW---GRSPSNKQTQPDQAQYGG 365
>UniRef100_Q9FX88 Putative RNA binding protein [Arabidopsis thaliana]
Length = 468
Score = 251 bits (641), Expect = 1e-65
Identities = 121/187 (64%), Positives = 147/187 (77%), Gaps = 2/187 (1%)
Query: 12 EEVRTLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAER 71
++V+TLW+GDL +W+DE YL CFSHT EV S+K+IRNK T Q EGYGF+EF+S SAAE
Sbjct: 116 DDVKTLWVGDLLHWMDETYLHTCFSHTNEVSSVKVIRNKQTCQSEGYGFVEFLSRSAAEE 175
Query: 72 VLQTYNGTQMPGTEQTFRLNWASFGIGERRP-DAGPDHSIFVGDLAPDVTDYLLQETFRT 130
LQ+++G MP EQ FRLNWASF GE+R + GPD SIFVGDLAPDV+D +L ETF
Sbjct: 176 ALQSFSGVTMPNAEQPFRLNWASFSTGEKRASENGPDLSIFVGDLAPDVSDAVLLETFAG 235
Query: 131 HYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKT 190
Y SV+GAKVV D NTGRSKGYGFV+F DE+ER+RAM+EMNG +CS+R MR+ ATPK+
Sbjct: 236 RYPSVKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMTEMNGAFCSSRQMRVGIATPKRA 295
Query: 191 TGY-QQN 196
Y QQN
Sbjct: 296 AAYGQQN 302
Score = 47.8 bits (112), Expect = 2e-04
Identities = 31/90 (34%), Positives = 46/90 (50%), Gaps = 8/90 (8%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V E L FS GEV+S+KI K G GF++F + +AE +
Sbjct: 351 TIFVGGLDADVTEEDLMQPFSDFGEVVSVKIPVGK------GCGFVQFANRQSAEEAIGN 404
Query: 76 YNGTQMPGTEQTFRLNWASFGIGERRPDAG 105
NGT + + T RL+W + R D+G
Sbjct: 405 LNGTVI--GKNTVRLSWGRSPNKQWRSDSG 432
Score = 47.4 bits (111), Expect = 3e-04
Identities = 50/212 (23%), Positives = 80/212 (37%), Gaps = 51/212 (24%)
Query: 16 TLWIGDLQYWVDENYLTHCFS-HTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L F+ V K++ + TG+ +GYGF+ F + R +
Sbjct: 214 SIFVGDLAPDVSDAVLLETFAGRYPSVKGAKVVIDSNTGRSKGYGFVRFGDENERSRAMT 273
Query: 75 TYNGT---------------------QMPGTEQTFR----LNWASF-------------- 95
NG Q G++ LN AS
Sbjct: 274 EMNGAFCSSRQMRVGIATPKRAAAYGQQNGSQGLITCLDALNIASEVNCNVFIGLALTLA 333
Query: 96 ----GIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPNTGRSKG 151
G G + +IFVG L DVT+ L + F + +G V K+ KG
Sbjct: 334 GGHGGNGSMSDGESNNSTIFVGGLDADVTEEDLMQPF-SDFGEVVSVKIPV------GKG 386
Query: 152 YGFVKFSDESERNRAMSEMNGVYCSTRPMRIS 183
GFV+F++ A+ +NG +R+S
Sbjct: 387 CGFVQFANRQSAEEAIGNLNGTVIGKNTVRLS 418
>UniRef100_Q9SX80 F16N3.23 protein [Arabidopsis thaliana]
Length = 434
Score = 244 bits (623), Expect = 1e-63
Identities = 125/217 (57%), Positives = 151/217 (68%), Gaps = 3/217 (1%)
Query: 6 HQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGE--VISIKIIRNKITGQPEGYGFIEF 63
HQ A E +T+W+GDLQ W+DE YL F+ E ++S+K+IRNK G EGYGF+EF
Sbjct: 94 HQNAFNGENKTIWVGDLQNWMDEAYLNSAFTSAEEREIVSLKVIRNKHNGSSEGYGFVEF 153
Query: 64 VSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERR-PDAGPDHSIFVGDLAPDVTDY 122
SH A++VLQ +NG MP T+Q FRLNWASF GE+R + GPD SIFVGDLAPDV+D
Sbjct: 154 ESHDVADKVLQEFNGAPMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLAPDVSDA 213
Query: 123 LLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRI 182
LL ETF Y SV+ AKVV D NTGRSKGYGFV+F DE+ER +AM+EMNGV CS+R MRI
Sbjct: 214 LLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRI 273
Query: 183 SAATPKKTTGYQQNPYAAVVAAAPVPKGITKPYTFFI 219
ATP+KT GYQQ A +G T T F+
Sbjct: 274 GPATPRKTNGYQQQGGYMPSGAFTRSEGDTINTTIFV 310
Score = 53.5 bits (127), Expect = 4e-06
Identities = 45/184 (24%), Positives = 81/184 (43%), Gaps = 25/184 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V + L FS V + K++ + TG+ +GYGF+ F + + +
Sbjct: 200 SIFVGDLAPDVSDALLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMT 259
Query: 75 TYNGTQMPGTEQTFRLNWAS---------------FGIGERRPDAGPDHSIFVGDLAPDV 119
NG + + + R+ A+ G R + +IFVG L V
Sbjct: 260 EMNGVKC--SSRAMRIGPATPRKTNGYQQQGGYMPSGAFTRSEGDTINTTIFVGGLDSSV 317
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
TD L++ F + +G + K+ KG GFV+F + A+ ++NG +
Sbjct: 318 TDEDLKQPF-SEFGEIVSVKIPV------GKGCGFVQFVNRPNAEEALEKLNGTVIGKQT 370
Query: 180 MRIS 183
+R+S
Sbjct: 371 VRLS 374
Score = 48.5 bits (114), Expect = 1e-04
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V + L FS GE++S+KI K G GF++FV+ AE L+
Sbjct: 307 TIFVGGLDSSVTDEDLKQPFSEFGEIVSVKIPVGK------GCGFVQFVNRPNAEEALEK 360
Query: 76 YNGTQMPGTEQTFRLNW 92
NGT + +QT RL+W
Sbjct: 361 LNGTVI--GKQTVRLSW 375
>UniRef100_Q9SX79 F16N3.24 protein [Arabidopsis thaliana]
Length = 432
Score = 242 bits (617), Expect = 6e-63
Identities = 122/217 (56%), Positives = 151/217 (69%), Gaps = 3/217 (1%)
Query: 6 HQPASLEEVRTLWIGDLQYWVDENYLTHCFSHTGE--VISIKIIRNKITGQPEGYGFIEF 63
HQ A E +T+W+GDL +W+DE YL F+ E ++S+K+IRNK G EGYGF+EF
Sbjct: 92 HQNAFNGENKTIWVGDLHHWMDEAYLNSSFASGDEREIVSVKVIRNKNNGLSEGYGFVEF 151
Query: 64 VSHSAAERVLQTYNGTQMPGTEQTFRLNWASFGIGERR-PDAGPDHSIFVGDLAPDVTDY 122
SH A++VL+ +NGT MP T+Q FRLNWASF GE+R + GPD SIFVGDL+PDV+D
Sbjct: 152 ESHDVADKVLREFNGTTMPNTDQPFRLNWASFSTGEKRLENNGPDLSIFVGDLSPDVSDN 211
Query: 123 LLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRI 182
LL ETF Y SV+ AKVV D NTGRSKGYGFV+F DE+ER +AM+EMNGV CS+R MRI
Sbjct: 212 LLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMTEMNGVKCSSRAMRI 271
Query: 183 SAATPKKTTGYQQNPYAAVVAAAPVPKGITKPYTFFI 219
ATP+KT GYQQ P+G T F+
Sbjct: 272 GPATPRKTNGYQQQGGYMPNGTLTRPEGDIMNTTIFV 308
Score = 55.8 bits (133), Expect = 8e-07
Identities = 47/184 (25%), Positives = 85/184 (45%), Gaps = 25/184 (13%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V +N L FS V + K++ + TG+ +GYGF+ F + + +
Sbjct: 198 SIFVGDLSPDVSDNLLHETFSEKYPSVKAAKVVLDANTGRSKGYGFVRFGDENERTKAMT 257
Query: 75 TYNGTQMPGTEQTFRLNWAS----FGIGER----------RPDAG-PDHSIFVGDLAPDV 119
NG + + + R+ A+ G ++ RP+ + +IFVG L V
Sbjct: 258 EMNGVKC--SSRAMRIGPATPRKTNGYQQQGGYMPNGTLTRPEGDIMNTTIFVGGLDSSV 315
Query: 120 TDYLLQETFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRP 179
TD L++ F +G + K+ KG GFV+F + A+ ++NG +
Sbjct: 316 TDEDLKQPFN-EFGEIVSVKIPV------GKGCGFVQFVNRPNAEEALEKLNGTVIGKQT 368
Query: 180 MRIS 183
+R+S
Sbjct: 369 VRLS 372
Score = 47.4 bits (111), Expect = 3e-04
Identities = 28/77 (36%), Positives = 43/77 (55%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V + L F+ GE++S+KI K G GF++FV+ AE L+
Sbjct: 305 TIFVGGLDSSVTDEDLKQPFNEFGEIVSVKIPVGK------GCGFVQFVNRPNAEEALEK 358
Query: 76 YNGTQMPGTEQTFRLNW 92
NGT + +QT RL+W
Sbjct: 359 LNGTVI--GKQTVRLSW 373
>UniRef100_Q69ME2 Putative nucleic acid binding protein [Oryza sativa]
Length = 316
Score = 239 bits (610), Expect = 4e-62
Identities = 112/167 (67%), Positives = 134/167 (80%)
Query: 26 VDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQTYNGTQMPGTE 85
+DENYL CF +TGEV++IK+IRNK TGQ EGYGF+EF SH+AAE+VL+ + G MP T+
Sbjct: 1 MDENYLHSCFGYTGEVVAIKVIRNKQTGQSEGYGFVEFYSHAAAEKVLEGFAGHIMPNTD 60
Query: 86 QTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQETFRTHYGSVRGAKVVTDPN 145
Q FR+NWASF +G+RR D DHSIFVGDLA DV D L ETF Y SV+GAKVV D N
Sbjct: 61 QPFRINWASFSMGDRRSDIASDHSIFVGDLASDVNDTTLLETFSKRYSSVKGAKVVIDAN 120
Query: 146 TGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRISAATPKKTTG 192
TGRSKGYGFV+F D++E+ AM+EMNGVYCSTRPMRI ATP+KT+G
Sbjct: 121 TGRSKGYGFVRFGDDNEKTHAMTEMNGVYCSTRPMRIGPATPRKTSG 167
Score = 55.5 bits (132), Expect = 1e-06
Identities = 44/177 (24%), Positives = 76/177 (42%), Gaps = 16/177 (9%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHT-GEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQ 74
++++GDL V++ L FS V K++ + TG+ +GYGF+ F + +
Sbjct: 84 SIFVGDLASDVNDTTLLETFSKRYSSVKGAKVVIDANTGRSKGYGFVRFGDDNEKTHAMT 143
Query: 75 TYNGTQM--------PGTEQTFRLNWASFGIGERRPDAGPDHSIFVGDLAPDVTDYLLQE 126
NG P T + G R + ++FVG L P+V++ L++
Sbjct: 144 EMNGVYCSTRPMRIGPATPRKTSGTSGPTGSAARSDGDLTNTTVFVGGLDPNVSEDDLRQ 203
Query: 127 TFRTHYGSVRGAKVVTDPNTGRSKGYGFVKFSDESERNRAMSEMNGVYCSTRPMRIS 183
TF + YG + K+ K GFV+F A+ +NG + +R+S
Sbjct: 204 TF-SQYGEISSVKIPV------GKQCGFVQFVQRKNAEDALQGLNGSTIGKQTVRLS 253
Score = 43.9 bits (102), Expect = 0.003
Identities = 29/77 (37%), Positives = 41/77 (52%), Gaps = 8/77 (10%)
Query: 16 TLWIGDLQYWVDENYLTHCFSHTGEVISIKIIRNKITGQPEGYGFIEFVSHSAAERVLQT 75
T+++G L V E+ L FS GE+ S+KI K GF++FV AE LQ
Sbjct: 186 TVFVGGLDPNVSEDDLRQTFSQYGEISSVKIPVGK------QCGFVQFVQRKNAEDALQG 239
Query: 76 YNGTQMPGTEQTFRLNW 92
NG+ + +QT RL+W
Sbjct: 240 LNGSTI--GKQTVRLSW 254
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.319 0.136 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 407,703,199
Number of Sequences: 2790947
Number of extensions: 16630459
Number of successful extensions: 47338
Number of sequences better than 10.0: 4111
Number of HSP's better than 10.0 without gapping: 2915
Number of HSP's successfully gapped in prelim test: 1198
Number of HSP's that attempted gapping in prelim test: 35530
Number of HSP's gapped (non-prelim): 8918
length of query: 230
length of database: 848,049,833
effective HSP length: 123
effective length of query: 107
effective length of database: 504,763,352
effective search space: 54009678664
effective search space used: 54009678664
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 72 (32.3 bits)
Medicago: description of AC146330.6


