

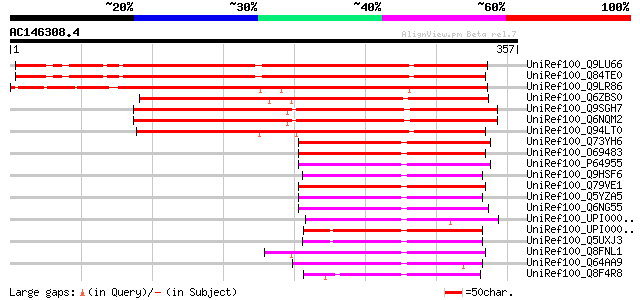
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC146308.4 + phase: 0
(357 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LU66 Similarity to RNase H [Arabidopsis thaliana] 310 5e-83
UniRef100_Q84TE0 At5g51080 [Arabidopsis thaliana] 307 3e-82
UniRef100_Q9LR86 T23E23.24 [Arabidopsis thaliana] 290 4e-77
UniRef100_Q6ZBS0 Putative RNase H domain-containing protein [Ory... 280 4e-74
UniRef100_Q9SGH7 Putative RNase H [Arabidopsis thaliana] 259 1e-67
UniRef100_Q6NQM2 At3g01410 [Arabidopsis thaliana] 258 2e-67
UniRef100_Q94LT0 Putative RNase [Oryza sativa] 250 4e-65
UniRef100_Q73YH6 Hypothetical protein [Mycobacterium paratubercu... 98 3e-19
UniRef100_O69483 Hypothetical protein MLCB1243.38 [Mycobacterium... 98 4e-19
UniRef100_P64955 Hypothetical protein Rv2228c/MT2287 [Mycobacter... 96 2e-18
UniRef100_Q9HSF6 Vng0255c [Halobacterium sp.] 92 3e-17
UniRef100_Q79VE1 Phosphoglycerate mutase/fructose-2,6-bisphospha... 91 6e-17
UniRef100_Q5YZA5 Putative phosphoglycerate mutase [Nocardia farc... 90 1e-16
UniRef100_Q6NG55 Hypothetical protein [Corynebacterium diphtheriae] 89 1e-16
UniRef100_UPI00002C6531 UPI00002C6531 UniRef100 entry 87 5e-16
UniRef100_UPI000033D4D0 UPI000033D4D0 UniRef100 entry 87 7e-16
UniRef100_Q5UXJ3 Ribonuclease H-like protein [Haloarcula marismo... 86 1e-15
UniRef100_Q8FNL1 Hypothetical protein [Corynebacterium efficiens] 86 2e-15
UniRef100_Q64AA9 Hypothetical protein [uncultured archaeon GZfos... 86 2e-15
UniRef100_Q8F4R8 Ribonuclease H-like protein [Leptospira interro... 83 1e-14
>UniRef100_Q9LU66 Similarity to RNase H [Arabidopsis thaliana]
Length = 316
Score = 310 bits (793), Expect = 5e-83
Identities = 170/332 (51%), Positives = 223/332 (66%), Gaps = 19/332 (5%)
Query: 5 FSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTRCYST 64
FS S+ S +L R + +V + P F + SL +S V + CYS+
Sbjct: 4 FSRARSYISLVLFRKSSYVTSH----IPWNQCF----YTSLKSSLKPASVSVSSVHCYSS 55
Query: 65 RKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPP 124
R KT SK+ K + +++ E DAF+VVRKGD+VGIY L D QAQVGSSV DPP
Sbjct: 56 RS--KTAKSKMSK--SSVSVSDSDKEKDAFFVVRKGDIVGIYKDLIDCQAQVGSSVYDPP 111
Query: 125 VSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNA 184
VSVYKGYS+ +TEE L + GLK LY RA DLKED+FG L PC FQD Q +++
Sbjct: 112 VSVYKGYSLLKDTEECLSTVGLKKPLYVFRALDLKEDMFGALTPCLFQD----QLPSASM 167
Query: 185 DSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATN 244
K LE + +TCI+EFDGASKGNPG +GA A+L+++DG+LI+++R+G+GIATN
Sbjct: 168 SVEKLAELEPSADTSYETCIIEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATN 227
Query: 245 NVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKEL 304
N AEY +ILG++HA++KG+T I ++ DSKLVCMQMK G WKV +E LS L+K AK+L
Sbjct: 228 NAAEYHGLILGLKHAIEKGYTKIKVKTDSKLVCMQMK---GQWKVNHEVLSKLHKEAKQL 284
Query: 305 KDKFVSFKISHVLRDLNSEADAQANLAVNLAG 336
DK +SF+ISHVLR LNS+AD QAN+A L+G
Sbjct: 285 SDKCLSFEISHVLRSLNSDADEQANMAARLSG 316
>UniRef100_Q84TE0 At5g51080 [Arabidopsis thaliana]
Length = 322
Score = 307 bits (787), Expect = 3e-82
Identities = 169/331 (51%), Positives = 222/331 (67%), Gaps = 19/331 (5%)
Query: 5 FSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTRCYST 64
FS S+ S +L R + +V + P F + SL +S V + CYS+
Sbjct: 4 FSRARSYISLVLFRKSSYVTSH----IPWNQCF----YTSLKSSLKPASVSVSSVHCYSS 55
Query: 65 RKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPP 124
R KT SK+ K + +++ E DAF+VVRKGD+VGIY L D QAQVGSSV DPP
Sbjct: 56 RS--KTAKSKMSK--SSVSVSDSDKEKDAFFVVRKGDIVGIYKDLIDCQAQVGSSVYDPP 111
Query: 125 VSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNA 184
VSVYKGYS+ +TEE L + GLK LY RA DLKED+FG L PC FQD Q +++
Sbjct: 112 VSVYKGYSLLKDTEECLSTVGLKKPLYVFRALDLKEDMFGALTPCLFQD----QLPSASM 167
Query: 185 DSSKKRALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATN 244
K LE + +TCI+EFDGASKGNPG +GA A+L+++DG+LI+++R+G+GIATN
Sbjct: 168 SVEKLAELEPSADTSYETCIIEFDGASKGNPGLSGAAAVLKTEDGSLIFKMRQGLGIATN 227
Query: 245 NVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKEL 304
N AEY +ILG++HA++KG+T I ++ DSKLVCMQMK G WKV +E LS L+K AK+L
Sbjct: 228 NAAEYHGLILGLKHAIEKGYTKIKVKTDSKLVCMQMK---GQWKVNHEVLSKLHKEAKQL 284
Query: 305 KDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
DK +SF+ISHVLR LNS+AD QAN+A L+
Sbjct: 285 SDKCLSFEISHVLRSLNSDADEQANMAARLS 315
>UniRef100_Q9LR86 T23E23.24 [Arabidopsis thaliana]
Length = 360
Score = 290 bits (742), Expect = 4e-77
Identities = 173/370 (46%), Positives = 236/370 (63%), Gaps = 44/370 (11%)
Query: 1 MNALFSHLSSFTSTILARTTQFVANRSLHTTPHTFSFPVKSHHSLLTSFHSQFVLTTVTR 60
MN L SH S+ + L + + +V+ S+ F P KS + + S F + +V
Sbjct: 1 MNCL-SHARSYIALGLLKRSSYVS--SIPWNECFFYMPSKSCLKPV-AVSSVFGICSVHS 56
Query: 61 CYSTRKLRKTGSSKLHKVEAETTMNQEQNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSV 120
S K K+ K+ + T ++ E DAF+VVRKGDV+GIY L+D QAQVGSSV
Sbjct: 57 YSSRSKAVKS------KMLSSTVVSAVDKEKDAFFVVRKGDVIGIYKDLSDCQAQVGSSV 110
Query: 121 CDPPVSVYKGYSMSNETEEYLLSHGLKNALYTIRASDLKEDLFGTLVPCPFQDPS----- 175
D PVSVYKGYS+ +TEEYL S GLK LY++RASDLK+D+FG L PC FQ+P+
Sbjct: 111 FDLPVSVYKGYSLPKDTEEYLSSVGLKKPLYSLRASDLKDDMFGALTPCLFQEPAPCTVK 170
Query: 176 ----STQGTTSNADSSKKR------ALEVLEQNNVKTCIVEFDGASKGNPGKAGAGAILR 225
T T + D K + + + LE+ + +TC +EFDGASKGNPG +GA A+L+
Sbjct: 171 VSEDETTSETKSKDDKKDQLPSASISYDPLEKLSKETCFIEFDGASKGNPGLSGAAAVLK 230
Query: 226 SKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQM----- 280
++DG+LI RVR+G+GIATNN AEY A+ILG+++A++KG+ +I ++GDSKLVCMQ+
Sbjct: 231 TEDGSLICRVRQGLGIATNNAAEYHALILGLKYAIEKGYKNIKVKGDSKLVCMQVSLMNH 290
Query: 281 --------------KQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
+QI G WKV +E L+ L+K AK L +K VSF+ISHVLR+LN++AD
Sbjct: 291 IRFYLLTFSTLSEKQQIKGQWKVNHEVLAKLHKEAKLLCNKCVSFEISHVLRNLNADADE 350
Query: 327 QANLAVNLAG 336
QANLAV L G
Sbjct: 351 QANLAVRLPG 360
>UniRef100_Q6ZBS0 Putative RNase H domain-containing protein [Oryza sativa]
Length = 351
Score = 280 bits (716), Expect = 4e-74
Identities = 141/255 (55%), Positives = 187/255 (73%), Gaps = 12/255 (4%)
Query: 92 DAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLKNALY 151
+ FYVVRKGDV+GIY +L+D QAQV +SVCDP V+VYKGYS+ ETEEYL + GL+N LY
Sbjct: 86 EPFYVVRKGDVIGIYKSLSDCQAQVSNSVCDPSVTVYKGYSLRKETEEYLAARGLRNPLY 145
Query: 152 TIRASDLKEDLFGTLVPCPFQDPSSTQGTT-----SNADSSKKRALEVLEQ----NNVKT 202
+I A+D +++LF LVPCPFQ P T +T K+ +V EQ N+ +
Sbjct: 146 SINAADARDELFDDLVPCPFQQPDGTGTSTLKRPLEMETGPSKKQPKVSEQEPLPNSSLS 205
Query: 203 CIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
C++EFDGASKGNPGKAGAGA++R DG +I ++REG+GIATNN AEYRA+ILG+ +A KK
Sbjct: 206 CLLEFDGASKGNPGKAGAGAVIRRLDGTVIAQLREGLGIATNNAAEYRALILGLTYAAKK 265
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
GF I QGDSKLVC Q+ +W+ +++ ++ L K KE+K +F +F+I+HVLR+ N+
Sbjct: 266 GFKYIRAQGDSKLVC---NQVSDVWRARHDTMADLCKRVKEIKGRFHTFQINHVLREFNT 322
Query: 323 EADAQANLAVNLAGE 337
+ADAQANLAV L GE
Sbjct: 323 DADAQANLAVELPGE 337
>UniRef100_Q9SGH7 Putative RNase H [Arabidopsis thaliana]
Length = 290
Score = 259 bits (661), Expect = 1e-67
Identities = 134/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G+K
Sbjct: 2 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIK 61
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 62 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 121
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 122 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 179
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 180 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 236
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 237 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 288
>UniRef100_Q6NQM2 At3g01410 [Arabidopsis thaliana]
Length = 294
Score = 258 bits (658), Expect = 2e-67
Identities = 133/292 (45%), Positives = 185/292 (62%), Gaps = 41/292 (14%)
Query: 88 QNESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLK 147
++E DAFY+VRKGD++G+Y +L++ Q Q GSSV P +SVYKGY E+ L S G++
Sbjct: 6 EDEKDAFYIVRKGDIIGVYRSLSECQGQAGSSVSHPAMSVYKGYGWPKGAEDLLSSFGIR 65
Query: 148 NALYTIRASDLKEDLFGTLVPCPFQDPSSTQGTTSNADSSKKRALEV------------- 194
NAL+++ AS +K+D FG L+PCP Q PSS+QG + N S KR ++
Sbjct: 66 NALFSVNASHVKDDAFGKLIPCPVQQPSSSQGESLNKSSPSKRLQDMGSGESGSFSPSPP 125
Query: 195 -----------------------LEQNNVKTCIVEFDGASKGNPGKAGAGAILRSKDGNL 231
+ QN+ +C +EFDGASKGNPGKAGAGA+LR+ D ++
Sbjct: 126 QKQLKIENDMLRRIPSSLLTRTPIRQND--SCTIEFDGASKGNPGKAGAGAVLRASDNSV 183
Query: 232 IYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKN 291
++ +REGVG ATNNVAEYRA++LG+R AL KGF ++ + GDS LVCM Q+ G WK +
Sbjct: 184 LFYLREGVGNATNNVAEYRALLLGLRSALDKGFKNVHVLGDSMLVCM---QVQGAWKTNH 240
Query: 292 ENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLAGEAFKIIN 343
++ L K AKEL + F +F I H+ R+ NSEAD QAN A+ LA ++I+
Sbjct: 241 PKMAELCKQAKELMNSFKTFDIKHIAREKNSEADKQANSAIFLADGQTQVIS 292
>UniRef100_Q94LT0 Putative RNase [Oryza sativa]
Length = 289
Score = 250 bits (639), Expect = 4e-65
Identities = 131/279 (46%), Positives = 176/279 (62%), Gaps = 36/279 (12%)
Query: 90 ESDAFYVVRKGDVVGIYNTLTDSQAQVGSSVCDPPVSVYKGYSMSNETEEYLLSHGLKNA 149
E +Y+VRKG++V +Y +L D QAQ+ SSV P S YKG S S E EEYL S GL NA
Sbjct: 3 EGSNYYLVRKGEMVAVYKSLNDCQAQICSSVSGPAASAYKGNSWSREKEEYLSSRGLSNA 62
Query: 150 LYTIRASDLKEDLFGTLVPCPFQDP------------------SSTQGTTSNADSSKKRA 191
Y I A++L+EDLFGTL+PC FQD S +G N S+ +
Sbjct: 63 TYVINAAELREDLFGTLIPCTFQDAVGSGQASAEHYSQRINQGYSVRGQAFNRLESRPSS 122
Query: 192 LEVLEQNNV---------------KTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVR 236
NN+ C++ FDGASKGNPGKAGAGA+L ++DG +I R+R
Sbjct: 123 SSHFSPNNLDQSGTVDAQPLSKQYMVCLLHFDGASKGNPGKAGAGAVLMTEDGRVISRLR 182
Query: 237 EGVGIATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLST 296
EG+GI TNNVAEYR +ILG+R+A++ GF I + GDS+LVC Q+K G W+ KN+N+
Sbjct: 183 EGLGIVTNNVAEYRGLILGLRYAIRHGFKKIIVYGDSQLVCYQVK---GTWQTKNQNMME 239
Query: 297 LYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
L K ++LK+ FVSF+I+H+ R+ N+EAD QAN+A+ L+
Sbjct: 240 LCKEVRKLKENFVSFEINHIRREWNAEADRQANIAITLS 278
>UniRef100_Q73YH6 Hypothetical protein [Mycobacterium paratuberculosis]
Length = 377
Score = 98.2 bits (243), Expect = 3e-19
Identities = 55/136 (40%), Positives = 84/136 (61%), Gaps = 4/136 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
++E DG S+GNPG AG GA++ + D G ++ ++ +G ATNNVAEYR ++ G+ ALK
Sbjct: 4 VIEADGGSRGNPGPAGYGAVVWTPDRGTVLAENKQAIGRATNNVAEYRGLLAGLGDALKL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKL+ ++Q+ G WKVK+ +L L+ A+ L +F + + R+ NS
Sbjct: 64 GATEAEVYLDSKLL---VEQMSGRWKVKHPDLIELHAQARSLAARFDRISYTWIPRERNS 120
Query: 323 EADAQANLAVNLAGEA 338
AD AN A++ A A
Sbjct: 121 HADRLANEAMDAAAAA 136
>UniRef100_O69483 Hypothetical protein MLCB1243.38 [Mycobacterium leprae]
Length = 371
Score = 97.8 bits (242), Expect = 4e-19
Identities = 57/133 (42%), Positives = 81/133 (60%), Gaps = 4/133 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDGNLIY-RVREGVGIATNNVAEYRAMILGMRHALKK 262
I+E DG S+GNPG AG GA++ D + + ++ +G ATNNVAEYRA+I G+ A+K
Sbjct: 4 IIEADGGSRGNPGPAGYGAVVWIADRSAVLTETKQAIGRATNNVAEYRALIAGLDDAVKM 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKLV ++Q+ G WKVK+ +L LY A+ L + S + + R NS
Sbjct: 64 GATEAEVLMDSKLV---VEQMSGRWKVKHPDLIELYVHAQTLASRLASVSYTWIPRTRNS 120
Query: 323 EADAQANLAVNLA 335
AD AN A++ A
Sbjct: 121 RADRLANEAMDAA 133
>UniRef100_P64955 Hypothetical protein Rv2228c/MT2287 [Mycobacterium tuberculosis]
Length = 364
Score = 95.9 bits (237), Expect = 2e-18
Identities = 55/136 (40%), Positives = 82/136 (59%), Gaps = 4/136 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
++E DG S+GNPG AG GA++ + D ++ ++ +G ATNNVAEYR +I G+ A+K
Sbjct: 4 VIEADGGSRGNPGPAGYGAVVWTADHSTVLAESKQAIGRATNNVAEYRGLIAGLDDAVKL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + DSKLV ++Q+ G WKVK+ +L LY A+ L +F V R N+
Sbjct: 64 GATEAAVLMDSKLV---VEQMSGRWKVKHPDLLKLYVQAQALASQFRRINYEWVPRARNT 120
Query: 323 EADAQANLAVNLAGEA 338
AD AN A++ A ++
Sbjct: 121 YADRLANDAMDAAAQS 136
>UniRef100_Q9HSF6 Vng0255c [Halobacterium sp.]
Length = 199
Score = 91.7 bits (226), Expect = 3e-17
Identities = 54/127 (42%), Positives = 73/127 (56%), Gaps = 3/127 (2%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTS 266
FDGAS+GNPG A G +L S DG ++ + +G ATNN AEY A+I + A GF
Sbjct: 74 FDGASRGNPGPAAVGWVLVSGDGGIVAEGGDTIGRATNNQAEYDALIAALEAAADFGFDD 133
Query: 267 ICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
I ++GDS+LV KQ+ G W + +L A+EL F + I+HV R N ADA
Sbjct: 134 IELRGDSQLV---EKQLTGAWDTNDPDLRRKRVRARELLTGFDDWSITHVPRATNERADA 190
Query: 327 QANLAVN 333
AN A++
Sbjct: 191 LANEALD 197
>UniRef100_Q79VE1 Phosphoglycerate mutase/fructose-2,6-bisphosphatase
[Corynebacterium glutamicum]
Length = 382
Score = 90.5 bits (223), Expect = 6e-17
Identities = 51/134 (38%), Positives = 84/134 (62%), Gaps = 5/134 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKD-GNLIYRVREGVGI-ATNNVAEYRAMILGMRHALK 261
++E DG S+GNPG AG+G ++ S + ++ + VG ATNNVAEYR ++ G++ A +
Sbjct: 4 VIEADGGSRGNPGVAGSGTVVYSDNKAEVLKEIAYVVGTKATNNVAEYRGLLEGLKAARE 63
Query: 262 KGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLN 321
G TS+ + DSKLV ++Q+ G WK+K+ ++ L AKE+ + S + + R+ N
Sbjct: 64 LGATSVDVYMDSKLV---VEQMSGRWKIKHPDMKVLAIEAKEIASEIGSVSYTWIPREKN 120
Query: 322 SEADAQANLAVNLA 335
ADA +N+A++ A
Sbjct: 121 KRADALSNVAMDAA 134
>UniRef100_Q5YZA5 Putative phosphoglycerate mutase [Nocardia farcinica]
Length = 406
Score = 89.7 bits (221), Expect = 1e-16
Identities = 55/131 (41%), Positives = 79/131 (59%), Gaps = 4/131 (3%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDG-NLIYRVREGVGIATNNVAEYRAMILGMRHALKK 262
IVE DG S+GNPG AG GA++ D ++ RE VGIATNNVAEYR +I G+ A +
Sbjct: 7 IVEADGGSRGNPGPAGYGAVVFDADHVAVLAERRESVGIATNNVAEYRGLIAGLEAAAEL 66
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G ++ ++ DSKLV ++Q+ G WKVK+ + L A+ L F + + + R N+
Sbjct: 67 GARTVDVRMDSKLV---VEQMSGRWKVKHAAMIPLADRARRLVAGFDAVTFTWIPRAQNA 123
Query: 323 EADAQANLAVN 333
AD AN A++
Sbjct: 124 HADRLANEAMD 134
>UniRef100_Q6NG55 Hypothetical protein [Corynebacterium diphtheriae]
Length = 377
Score = 89.4 bits (220), Expect = 1e-16
Identities = 51/135 (37%), Positives = 79/135 (57%), Gaps = 4/135 (2%)
Query: 204 IVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGV-GIATNNVAEYRAMILGMRHALKK 262
I+E DG S+GNPG AG+G ++ + D + + + V G ATNNVAEY ++ G+R A +
Sbjct: 4 IIEADGGSRGNPGIAGSGTLIYNADRSQVLKEYSYVVGKATNNVAEYHGLLNGLRGAAEL 63
Query: 263 GFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNS 322
G T + ++ DSKLV ++Q+ G WK+K+ ++ L K++ S + R NS
Sbjct: 64 GATEVSVRMDSKLV---VEQMSGRWKIKHPDMKELALECKKVASSIGSVSYEWIPRSENS 120
Query: 323 EADAQANLAVNLAGE 337
ADA AN A++ E
Sbjct: 121 HADALANKAMDALAE 135
>UniRef100_UPI00002C6531 UPI00002C6531 UniRef100 entry
Length = 165
Score = 87.4 bits (215), Expect = 5e-16
Identities = 56/141 (39%), Positives = 82/141 (57%), Gaps = 8/141 (5%)
Query: 209 GASKGNPGKAGAGAIL-RSKDGNLIYRVREGVGIA-TNNVAEYRAMILGMRHALKKGFT- 265
GA +GNPG +GAGA+L R D +++ + E +G TNN AEY A+ G+R A G T
Sbjct: 1 GACRGNPGPSGAGALLRRDADDAIVFELAEYLGDRWTNNEAEYAALTRGLRVAADMGVTK 60
Query: 266 SICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFV--SFKISHVLRDLNSE 323
+ ++GDS LV + Q+ G WKVK NL+ L++ A+ + D I+HV R+ N E
Sbjct: 61 DLRVRGDSALV---VNQVAGTWKVKRPNLAPLHEDARAIIDAHFGGEIDIAHVDRERNRE 117
Query: 324 ADAQANLAVNLAGEAFKIINV 344
ADA AN + + + +NV
Sbjct: 118 ADALANAGITVGASVGEGVNV 138
>UniRef100_UPI000033D4D0 UPI000033D4D0 UniRef100 entry
Length = 130
Score = 87.0 bits (214), Expect = 7e-16
Identities = 51/126 (40%), Positives = 77/126 (60%), Gaps = 4/126 (3%)
Query: 208 DGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTSI 267
DGAS+ NPG+A G + + DG I+ + + +GIA+NN AEY A+ + + LK F ++
Sbjct: 8 DGASRSNPGEAAIGVSIET-DGKEIHTISKTIGIASNNEAEYLALDRALEYCLKNDFLNL 66
Query: 268 CIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQ 327
I DSKLV ++Q++G +KVK+ NL L E +KF I+H+ R+ N AD
Sbjct: 67 EIFLDSKLV---VEQVNGNFKVKSNNLKPLRDKILEHIEKFEFVSINHIYRENNKRADEL 123
Query: 328 ANLAVN 333
ANLA++
Sbjct: 124 ANLALD 129
>UniRef100_Q5UXJ3 Ribonuclease H-like protein [Haloarcula marismortui]
Length = 198
Score = 86.3 bits (212), Expect = 1e-15
Identities = 50/127 (39%), Positives = 73/127 (57%), Gaps = 4/127 (3%)
Query: 207 FDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFTS 266
FDGAS+GNPG A G +L + D ++ E +G ATNN AEY+A+I + A GF
Sbjct: 74 FDGASRGNPGPASVGYVLVN-DSGIVTEGGETIGTATNNQAEYKALIRAVEVARDYGFDD 132
Query: 267 ICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEADA 326
+ I+GDS+L+ +KQ+ G W + L +EL F ++I HV R++N AD
Sbjct: 133 VHIRGDSELI---VKQVRGEWDTNDPELREHRVRVRELLTDFDDWQIEHVPREINDRADE 189
Query: 327 QANLAVN 333
AN A++
Sbjct: 190 LANDALD 196
>UniRef100_Q8FNL1 Hypothetical protein [Corynebacterium efficiens]
Length = 435
Score = 85.9 bits (211), Expect = 2e-15
Identities = 58/162 (35%), Positives = 93/162 (56%), Gaps = 9/162 (5%)
Query: 180 TTSNADSSKKRALEVLEQ---NNVKT-CIVEFDGASKGNPGKAGAGAILRSKDGNLIYR- 234
TT+ K+R+ + + N+V I+E DG S+GNPG AG+G ++ S++ I R
Sbjct: 20 TTTGGGLRKRRSCQPVHDFYGNDVSMKLIIEADGGSRGNPGVAGSGTVVYSENRERILRE 79
Query: 235 VREGVGI-ATNNVAEYRAMILGMRHALKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNEN 293
+ VG +TNNVAEYR +I G+R A + G T + + DSKLV ++Q+ G WK+K+ +
Sbjct: 80 IAYVVGTRSTNNVAEYRGLIEGLRVARELGATEVEVFMDSKLV---VEQMSGRWKIKHPD 136
Query: 294 LSTLYKVAKELKDKFVSFKISHVLRDLNSEADAQANLAVNLA 335
+ TL A+ L + + R+ N AD +N+A++ A
Sbjct: 137 MKTLAMEARNLAGDIGRVTYTWIPREKNKAADHLSNVAMDAA 178
>UniRef100_Q64AA9 Hypothetical protein [uncultured archaeon GZfos32E7]
Length = 173
Score = 85.5 bits (210), Expect = 2e-15
Identities = 51/136 (37%), Positives = 79/136 (57%), Gaps = 5/136 (3%)
Query: 200 VKTCIVEFDGASKGNPGKAGAGAILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHA 259
+K I+ DGA +GNPG AG G ++ ++ G I +E +G ATNN+AEYRA+I + A
Sbjct: 15 MKKLIIYTDGACRGNPGPAGIGIVICNESGKKIKEDKEFIGDATNNIAEYRALIKALELA 74
Query: 260 LKKGFTSICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLR- 318
T + DS+L+ ++Q++G ++VK+E L L+ KE + F SHV R
Sbjct: 75 SDFSVTRVECFSDSELM---VRQLNGAYRVKDEKLGELFLQVKEKERLFEEVTYSHVPRK 131
Query: 319 -DLNSEADAQANLAVN 333
+L AD+ ANL ++
Sbjct: 132 NNLIKRADSLANLGID 147
>UniRef100_Q8F4R8 Ribonuclease H-like protein [Leptospira interrogans]
Length = 129
Score = 83.2 bits (204), Expect = 1e-14
Identities = 53/126 (42%), Positives = 74/126 (58%), Gaps = 8/126 (6%)
Query: 208 DGASKGNPGKAGAG--AILRSKDGNLIYRVREGVGIATNNVAEYRAMILGMRHALKKGFT 265
DGASKGNPG + G A + K+ +R+ E +G TNNVAE+ A+ G+ + + F
Sbjct: 7 DGASKGNPGPSSIGIVAYIHEKEE---FRISERIGETTNNVAEWSALKKGIEECISRKFD 63
Query: 266 SICIQGDSKLVCMQMKQIDGLWKVKNENLSTLYKVAKELKDKFVSFKISHVLRDLNSEAD 325
SI DS+LV +KQ++G +KVK+ NL K +L SF+I+HV R+ NS AD
Sbjct: 64 SIHAYMDSELV---VKQVNGKYKVKHPNLLEYKKEVDKLISSLHSFQITHVPREKNSVAD 120
Query: 326 AQANLA 331
AN A
Sbjct: 121 KLANEA 126
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.317 0.131 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 551,797,825
Number of Sequences: 2790947
Number of extensions: 22167687
Number of successful extensions: 65038
Number of sequences better than 10.0: 769
Number of HSP's better than 10.0 without gapping: 437
Number of HSP's successfully gapped in prelim test: 332
Number of HSP's that attempted gapping in prelim test: 64247
Number of HSP's gapped (non-prelim): 834
length of query: 357
length of database: 848,049,833
effective HSP length: 128
effective length of query: 229
effective length of database: 490,808,617
effective search space: 112395173293
effective search space used: 112395173293
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 75 (33.5 bits)
Medicago: description of AC146308.4


