

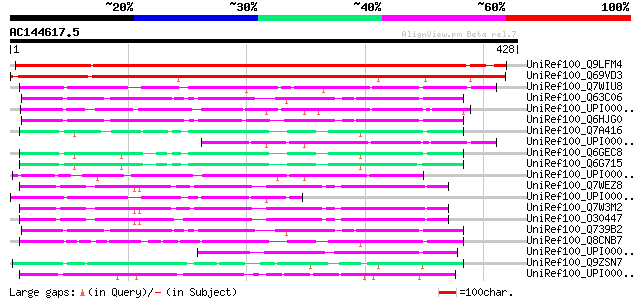
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.5 + phase: 0
(428 letters)
Database: uniref100
2,790,947 sequences; 848,049,833 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UniRef100_Q9LFM4 Hypothetical protein F2I11_220 [Arabidopsis tha... 549 e-155
UniRef100_Q69VD3 Monooxygenase-like [Oryza sativa] 460 e-128
UniRef100_Q7WIU8 Putative monooxygenase [Bordetella bronchiseptica] 107 8e-22
UniRef100_Q63C06 Probable FAD-dependent monooxygenase [Bacillus ... 96 1e-18
UniRef100_UPI00002A73B2 UPI00002A73B2 UniRef100 entry 94 7e-18
UniRef100_Q6HJG0 Possible FAD-dependent monooxygenase [Bacillus ... 94 7e-18
UniRef100_Q7A416 SA2099 protein [Staphylococcus aureus] 92 3e-17
UniRef100_UPI0000286020 UPI0000286020 UniRef100 entry 91 5e-17
UniRef100_Q6GEC8 Putative monooxygenase [Staphylococcus aureus] 91 5e-17
UniRef100_Q6G715 Putative monooxygenase [Staphylococcus aureus] 91 5e-17
UniRef100_UPI00003184E9 UPI00003184E9 UniRef100 entry 90 1e-16
UniRef100_Q7WEZ8 Putative hydroxylase [Bordetella bronchiseptica] 89 2e-16
UniRef100_UPI000031B0F6 UPI000031B0F6 UniRef100 entry 88 5e-16
UniRef100_Q7W3M2 Putative hydroxylase [Bordetella parapertussis] 87 9e-16
UniRef100_O30447 Hypothetical protein [Bordetella pertussis] 86 2e-15
UniRef100_Q739B2 FAD binding-monooxygenase family protein [Bacil... 81 5e-14
UniRef100_Q8CNB7 Monooxygenase [Staphylococcus epidermidis] 80 9e-14
UniRef100_UPI00002F808F UPI00002F808F UniRef100 entry 78 6e-13
UniRef100_Q9ZSN7 CTF2B [Arabidopsis thaliana] 77 9e-13
UniRef100_UPI00002FAF2D UPI00002FAF2D UniRef100 entry 76 2e-12
>UniRef100_Q9LFM4 Hypothetical protein F2I11_220 [Arabidopsis thaliana]
Length = 408
Score = 549 bits (1415), Expect = e-155
Identities = 275/414 (66%), Positives = 321/414 (77%), Gaps = 10/414 (2%)
Query: 6 RKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW 65
+K KA+IVGGSI G+S AH+L LA WDVLVLEK++ PP GSPTGAGLGL+ ++QII+SW
Sbjct: 4 KKGKAIIVGGSIAGLSCAHSLTLACWDVLVLEKSSEPPVGSPTGAGLGLDPQARQIIKSW 63
Query: 66 ISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQ 125
++ P Q L T PL+IDQN TDSEKKV R LTRDE +FRAA+W D+ +LY +LP
Sbjct: 64 LTTP-QLLDEITLPLSIDQNQTTDSEKKVTRILTRDEDFDFRAAYWSDIQSLLYNALPQT 122
Query: 126 IFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKL 185
+FLWGH FLSF ++ ++ V VK VV+T E +EI GDLL+AADGCLSSIR+ +LPDFKL
Sbjct: 123 MFLWGHKFLSFTMSQDESNVKVKTLVVETQETVEIEGDLLIAADGCLSSIRKTFLPDFKL 182
Query: 186 RYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNWI 245
RYSGYCAWRGV DFS ENSET+ GIKK YPDLGKCLYFDL THSV YEL NKKLNWI
Sbjct: 183 RYSGYCAWRGVFDFSGNENSETVSGIKKVYPDLGKCLYFDLGEQTHSVFYELFNKKLNWI 242
Query: 246 WYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPL 305
WYVNQPEP++K SVT KV+ +MI KMHQEAE IWIPELA++M ETK+PFLN IYDSDPL
Sbjct: 243 WYVNQPEPDLKSNSVTLKVSQEMINKMHQEAETIWIPELARLMSETKDPFLNVIYDSDPL 302
Query: 306 EKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRL 365
E+IFW N+VLVG+AAHPTTPH LRSTNM+ILDA VLGKC+E G E + LEEYQ RL
Sbjct: 303 ERIFWGNIVLVGDAAHPTTPHGLRSTNMSILDAEVLGKCLENCGPENVSLGLEEYQRIRL 362
Query: 366 PVTSKQVLHARRLGRIKQGLVLPDRDPFDPKSARQEECQEIILTNTPFFNDAPL 419
PV S+QVL+ARRLGRIKQGL D D E+ N PFF+ APL
Sbjct: 363 PVVSEQVLYARRLGRIKQGL---DHDGIGDGVFGLEQ------RNMPFFSCAPL 407
>UniRef100_Q69VD3 Monooxygenase-like [Oryza sativa]
Length = 431
Score = 460 bits (1184), Expect = e-128
Identities = 234/434 (53%), Positives = 303/434 (68%), Gaps = 22/434 (5%)
Query: 1 MVGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQ 60
MVG K AV+VGGS+ G++ AHA+ AGW+ +V+EK +P GS TGAGLGL++ S +
Sbjct: 1 MVG---KRVAVVVGGSVAGLACAHAVAEAGWEAVVVEKAAAPGAGSGTGAGLGLDAQSME 57
Query: 61 IIQSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYE 120
+ WI P L T PL +D N TD E K RTLTRDE FRAAHWGDLH L+E
Sbjct: 58 TLARWI---PGRLDAATLPLAVDLNRATDGETKAGRTLTRDEGFGFRAAHWGDLHRRLHE 114
Query: 121 SLPPQI-FLWGHIFLSFHVANE---KGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIR 176
+LP + LWGH F+SF +A E G V+V A+V++TGE +E+ GDLLVAADGC S+IR
Sbjct: 115 ALPAGVTVLWGHQFVSFEMAPEGDGDGGVVVTARVLRTGETVEVAGDLLVAADGCTSAIR 174
Query: 177 RKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYE 236
R++LP+ KLRYSGYCAWRGV DFS E T+ I++AYP+LG CLYFDLA TH+VLYE
Sbjct: 175 RRFLPELKLRYSGYCAWRGVFDFSGKEGCTTMVDIRRAYPELGNCLYFDLAYKTHAVLYE 234
Query: 237 LPNKKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFL 296
LP +LNW+WY+N EPE+ G+SVT KV + +M +EAE++W PELA+++ ET EPF+
Sbjct: 235 LPKNRLNWLWYINGDEPELMGSSVTMKVNEATVSEMKEEAERVWCPELARLIGETAEPFV 294
Query: 297 NFIYDSDPLEKIFW--DNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWG----- 349
N IYD++PL + W V LVG+AAHPTTPH LRSTNM+ILDA VLG C+ +WG
Sbjct: 295 NVIYDAEPLPGLSWAGGRVALVGDAAHPTTPHGLRSTNMSILDARVLGCCLARWGGDGNA 354
Query: 350 --TEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVL---PDRDPFDPKSARQEECQ 404
T AL EY+ R PV + QVLHARRLGR+KQGL + D + FD ++A +EE
Sbjct: 355 EPTSTPRRALAEYEAARRPVVAAQVLHARRLGRLKQGLGMGSAGDGEGFDARTATEEEIS 414
Query: 405 EIILTNTPFFNDAP 418
++ ++ P+F+ AP
Sbjct: 415 QLRQSSMPYFSGAP 428
>UniRef100_Q7WIU8 Putative monooxygenase [Bordetella bronchiseptica]
Length = 402
Score = 107 bits (266), Expect = 8e-22
Identities = 105/420 (25%), Positives = 179/420 (42%), Gaps = 44/420 (10%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
KA ++GGS+GG+ +A+ L+ AGWDV V E+ G GAG+ + +++
Sbjct: 3 KAAVIGGSLGGLFAANLLLRAGWDVHVHERVHDELDGR--GAGIVTHPELLDVLRRIGVA 60
Query: 69 PPQFLH-NTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIF 127
P + + +T+D + + + + + LT WG L +L P + +
Sbjct: 61 PTESIGIEVARRVTLDLDGSVLASRALPQLLTA----------WGRLFSILRTRFPAERY 110
Query: 128 LWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRY 187
G S + +++ + DL+VAADG S++R+ LP + Y
Sbjct: 111 HRGQALESLEQDERRA-------LLRFADGARAEVDLVVAADGLRSTLRQALLPQARPAY 163
Query: 188 SGYCAWRGVLD---FSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNW 244
+GY AWRG++D SE E P + + + +A GT S + ++ N+
Sbjct: 164 AGYVAWRGLVDEASLSERTRRELCPYFAFGLPAHEQMIAYPVA-GTGSGAQDAA-RRFNF 221
Query: 245 IWYVNQPEPEVKGTSVTTK-------------VTSDMIQKMHQEAEKIWIPELAKVMKET 291
+WY E VT + +++ + Q A ++ P+ +V+ +T
Sbjct: 222 VWYRPADEHTTFRDMVTDAQGQAWMDGIPPPLIRPEIVAQARQAAAQVLAPQFGEVVAKT 281
Query: 292 KEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTE 351
F I+D + ++ V L+G+AA PH DA L + GT
Sbjct: 282 ANLFFQPIFDLES-PRLALGRVALLGDAAFVARPHCGMGVTKAAGDARALAYALA--GTP 338
Query: 352 MLESALEEYQLTRLPVTSKQVLHARRLGRIKQGLVLPDRDPFDPKSARQEECQEIILTNT 411
+ +AL+ Y+ RLP V HAR LGR Q + RD + + A + E I+ T
Sbjct: 339 DIGAALQAYEDERLPFGKFIVGHARDLGRYMQAQL---RDDAEREMAERYRTPEAIMRET 395
>UniRef100_Q63C06 Probable FAD-dependent monooxygenase [Bacillus cereus]
Length = 377
Score = 96.3 bits (238), Expect = 1e-18
Identities = 96/376 (25%), Positives = 159/376 (41%), Gaps = 40/376 (10%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-ISKP 69
+I+GG I G+ +A +L G DV V +K T P GAG+ + + Q ++ + ISK
Sbjct: 5 IIIGGGIAGLCAAISLQKIGLDVKVYDKNTEPTVA---GAGIIIAPNAMQALELYGISKK 61
Query: 70 PQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLW 129
+ N ++ NLV++ N+ + + H DLH +L L W
Sbjct: 62 IKKFGNESNGF----NLVSEKGTTFNKLIIPTCYPKMYSIHRKDLHQLLLSELKEDTVKW 117
Query: 130 GHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSG 189
G + NE + + V + G E +G++L+AADG S IR++ RY+G
Sbjct: 118 GKECVKIE-QNEANALKI---VFQDGS--EALGNILIAADGIHSVIRKQVTQGDNYRYAG 171
Query: 190 YCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHS--VLYELPNKKLNWIWY 247
Y WRGV + + + F GT+ + LPN ++ W
Sbjct: 172 YTCWRGVTPANNLSLTND----------------FIETWGTNGRFGIVPLPNNEVYWYAL 215
Query: 248 VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEK 307
+N + K + TT + + H IP + K + + I D P+++
Sbjct: 216 INAKARDPKYKAYTTTDLYNHFKTYHNP-----IPSILKNASDI-DMIHRDIIDITPMKQ 269
Query: 308 IFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPV 367
F +V +G+AAH TP+ + I DA +L +CI+ A EY+ R
Sbjct: 270 FFDKRIVFIGDAAHALTPNLGQGACQAIEDAIILAECIK--NNAHYRQAFLEYEQKRRDR 327
Query: 368 TSKQVLHARRLGRIKQ 383
K A ++G++ Q
Sbjct: 328 IEKISNTAWKVGKMAQ 343
>UniRef100_UPI00002A73B2 UPI00002A73B2 UniRef100 entry
Length = 399
Score = 94.0 bits (232), Expect = 7e-18
Identities = 93/403 (23%), Positives = 177/403 (43%), Gaps = 49/403 (12%)
Query: 10 AVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-LNSLSQQIIQSWISK 68
A I+GGS+ G+ +A+AL GWD+ + EK P +G GAG+ + L+ + +S
Sbjct: 5 ATIIGGSMAGLFAANALRKKGWDISIHEKVPVPLSG--RGAGIATYDELADLVFKS---- 58
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFL 128
+N + + D + K+ + + W L +L E + + +
Sbjct: 59 ---TGNNNVLGTSAKSRISLDQKGKIIASYDYPQVY----TSWQYLFSILREKIDNKNYF 111
Query: 129 WGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYS 188
G H EK + K K DL++ A+G S +R +Y+
Sbjct: 112 MGDDCTEIHHGKEKVTAVFNNKKEKQ-------SDLVIIANGIKSELRHYVDSTASPQYA 164
Query: 189 GYCAWRGVLDFSEIENSETIKGIKKAY----PDLGKCLYFDLASGTHSVLYELPNKKLNW 244
GY WRGV+ + + + ++++ + + P+ + + +A G +++ N+++NW
Sbjct: 165 GYVGWRGVV-YEDQLSKKSLEMLSNYFIVVLPNNQQIAAYPIA-GEGKDPFKIGNRRINW 222
Query: 245 IWY----VNQPEPEVKGTS--------VTTKVTSDMIQKMHQEAEKIWIPELAKVMKETK 292
IWY + + + G S ++ +++Q + +EAE P+L +++ +T
Sbjct: 223 IWYKPANTKKLDTLLLGKSGQQFVDGIPPNEIRDEIVQDLFKEAENKLPPQLTELVLKTS 282
Query: 293 EPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEM 352
+P + I+D + +K+ V+ +G+AA PH LDA L ++ G
Sbjct: 283 QPLIQPIFDLES-DKMLNKRVITIGDAAFTARPHIGMGVTKAALDAFTLSDHLQNKG--Y 339
Query: 353 LESALEEYQLTRLPVTSKQVLHARRLGRI------KQGLVLPD 389
LE L ++ +R+ V +R LGR + LV+PD
Sbjct: 340 LEE-LINWEASRVESGKFLVNRSRLLGRYLSEPEKSETLVMPD 381
>UniRef100_Q6HJG0 Possible FAD-dependent monooxygenase [Bacillus thuringiensis]
Length = 377
Score = 94.0 bits (232), Expect = 7e-18
Identities = 93/374 (24%), Positives = 155/374 (40%), Gaps = 36/374 (9%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-ISKP 69
+I+GG I G+ +A +L G DV V +K T P GAG+ + + Q ++ + ISK
Sbjct: 5 IIIGGGIAGLCAAISLQKIGLDVKVYDKNTEPTVA---GAGIIIAPNAMQALELYGISKK 61
Query: 70 PQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLW 129
+ N + NLV+ N+ + + H DLH +L L W
Sbjct: 62 IKKFGNESDGF----NLVSKKGTTFNKLIIPTCYPKMYSIHRKDLHQLLLSELKEDTVKW 117
Query: 130 GHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSG 189
G + E IV + G E +G++L+AADG S +R++ RY+G
Sbjct: 118 GKECVKIEQNEENALKIV----FQDGS--EALGNILIAADGIHSIVRKQVTQGDNYRYAG 171
Query: 190 YCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNWIWYVN 249
Y WRGV + +N + + G+ + LPN ++ W +N
Sbjct: 172 YTCWRGV---TPTKNLSLTNDFIETWGTNGR-----------FGIVPLPNNEVYWYALIN 217
Query: 250 QPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIF 309
+ K + TT + + H IP + + + + I D P+++ F
Sbjct: 218 AKARDPKYKAYTTTDLYNHFKTYHNP-----IPSILQNASDI-DMIHRDIIDITPMKQFF 271
Query: 310 WDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTS 369
+V +G+AAH TP+ + I DA +L +CI+ A E++ R
Sbjct: 272 DKRIVFIGDAAHALTPNLGQGACQAIEDAIILAECIK--NNAYYRQAFTEFEQKRRDRIE 329
Query: 370 KQVLHARRLGRIKQ 383
K A ++G+I Q
Sbjct: 330 KISNTAWKVGKIAQ 343
>UniRef100_Q7A416 SA2099 protein [Staphylococcus aureus]
Length = 374
Score = 92.0 bits (227), Expect = 3e-17
Identities = 93/383 (24%), Positives = 154/383 (39%), Gaps = 53/383 (13%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLP 123
I Q L T ++ D ++ + + +LN L ++ +
Sbjct: 59 KGIKNAGQILSTMT--------VLDDKDRLLTTVKLKSNTLNVTLPRQ-TLIDIIKSYVK 109
Query: 124 PQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDF 183
+ H H+ NE V + E DL + ADG S +R+ D
Sbjct: 110 DDVIFTNHEVT--HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNADS 162
Query: 184 KLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLN 243
K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 163 KVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQAY 208
Query: 244 WIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNFIY 300
W +N E K +S K H +A P E+ +++ + E L+ IY
Sbjct: 209 WFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHNIY 259
Query: 301 DSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEY 360
D PL+ + +L+G+AAH TTP+ + + DA VL C + E AL+ Y
Sbjct: 260 DLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNCFNAYD---FEKALQRY 316
Query: 361 QLTRLPVTSKQVLHARRLGRIKQ 383
R+ T+K + +R++G+I Q
Sbjct: 317 DKIRVKHTAKVIKRSRKIGKIAQ 339
>UniRef100_UPI0000286020 UPI0000286020 UniRef100 entry
Length = 286
Score = 91.3 bits (225), Expect = 5e-17
Identities = 71/266 (26%), Positives = 128/266 (47%), Gaps = 26/266 (9%)
Query: 163 DLLVAADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAY----PDL 218
DL++AADG SSIR ++LPD KL+Y+GY AWRG++D + + + ET + + + + P
Sbjct: 24 DLVIAADGFKSSIREQFLPDVKLQYAGYVAWRGLVDEAAL-SRETREALFEKFAFCLPPR 82
Query: 219 GKCLYFDLASGTHSVLYELPNKKLNWIWY-------------VNQPEPEVKGTSVTTKVT 265
+ L + +A +S ++ N++WY +Q G T +
Sbjct: 83 EQILGYPVAGQGNST--TPGERRYNFVWYRATSEDIQLPNLLTDQSGKRWVGGIPPTLIR 140
Query: 266 SDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTP 325
D++ M + A + P+ +V+ + K+P I+D + + ++ + + L+G+AA P
Sbjct: 141 QDILADMEEAALTLLAPQFGEVVTKAKQPLFQPIFDLE-VPQMAFGRIALLGDAAFVARP 199
Query: 326 HYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQGL 385
H DA L K ++ T++ E AL +Y R V + V HAR LG Q
Sbjct: 200 HCGMGVTKAASDAMALVKALQA-RTDVRE-ALADYSRERTQVGAAIVQHARHLGAYMQAQ 257
Query: 386 VLPDRDPFDPKSARQEECQEIILTNT 411
+ ++ D + A + + ++ T
Sbjct: 258 L---KNEVDREMAERYRTPDAVMRET 280
>UniRef100_Q6GEC8 Putative monooxygenase [Staphylococcus aureus]
Length = 374
Score = 91.3 bits (225), Expect = 5e-17
Identities = 99/385 (25%), Positives = 156/385 (39%), Gaps = 57/385 (14%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKK--VNRTLTRDESLNFRAAHWGDLHGVLYES 121
I Q L T D+ L T K +N TL R ++ ++ D
Sbjct: 59 KGIKNAGQILSTMTVLDDKDRPLTTVKLKSNTLNVTLPRQTLIDIIKSYVKD-------- 110
Query: 122 LPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLP 181
IF + H+ NE V + E DL + ADG S +R+
Sbjct: 111 --DAIFTNHEVT---HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNA 160
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
D K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 161 DSKVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQ 206
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNF 298
W +N E K +S K H +A P E+ +++ + E L+
Sbjct: 207 AYWFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHN 257
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
IYD PL+ + +L+G+AAH TTP+ + + DA VL C + E AL+
Sbjct: 258 IYDLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNCFNAYD---FEKALQ 314
Query: 359 EYQLTRLPVTSKQVLHARRLGRIKQ 383
Y R+ T+K + +R++G+I Q
Sbjct: 315 RYDKIRVKHTAKVIKRSRKIGKIAQ 339
>UniRef100_Q6G715 Putative monooxygenase [Staphylococcus aureus]
Length = 374
Score = 91.3 bits (225), Expect = 5e-17
Identities = 100/385 (25%), Positives = 157/385 (39%), Gaps = 57/385 (14%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLG-----LNSLSQQIIQ 63
K I+G IGG+++A L G + V EK S GAG+G L L +
Sbjct: 2 KIAIIGAGIGGLTAAALLQEQGHTIKVFEKNESV---KEIGAGIGIGDNVLKKLGNHDLA 58
Query: 64 SWISKPPQFLHNTTHPLTIDQNLVTDSEKK--VNRTLTRDESLNFRAAHWGDLHGVLYES 121
I Q L T D+ L T K +N TL R ++ ++ D
Sbjct: 59 KGIKNAGQILSTMTVLDDKDRLLTTVKLKSNTLNVTLPRQTLIDIIKSYVKD-------- 110
Query: 122 LPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLP 181
IF + H+ NE V + E DL + ADG S +R+
Sbjct: 111 --DAIFTNHEVT---HIDNETDKV-----TIHFAEQESEAFDLCIGADGIHSKVRQSVNA 160
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
D K+ Y GY +RG++D ++++ + K +G + L N +
Sbjct: 161 DSKVLYQGYTCFRGLIDDIDLKHPDCAKEYWGRKGRVG--------------IVPLLNNQ 206
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNF 298
W +N E K +S K H +A P E+ +++ + E L+
Sbjct: 207 AYWFITINSKENNHKYSS---------FGKPHLQAYFNHYPNEVREILDKQSETGILLHN 257
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
IYD PL+ + +L+G+AAH TTP+ + + DA VL C + T E AL+
Sbjct: 258 IYDLKPLKSFVYGRTILLGDAAHATTPNMGQGAGQAMEDAIVLVNC---FNTYDFEKALQ 314
Query: 359 EYQLTRLPVTSKQVLHARRLGRIKQ 383
Y R+ T+K + +R++G+I Q
Sbjct: 315 RYDKIRVKHTAKVIKRSRKIGKIAQ 339
>UniRef100_UPI00003184E9 UPI00003184E9 UniRef100 entry
Length = 494
Score = 89.7 bits (221), Expect = 1e-16
Identities = 96/365 (26%), Positives = 151/365 (41%), Gaps = 44/365 (12%)
Query: 3 GEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQII 62
G GRK AVIVGGS+GG+ +A AL AGWDV V E+ SP G G+ ++
Sbjct: 118 GSGRK--AVIVGGSVGGLFTATALRAAGWDVGVFEQ--SPHDLDSRGGGI--------VL 165
Query: 63 QSWISKPPQF----LHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVL 118
Q I + F L +ID+ V ++ V R ++ W ++ L
Sbjct: 166 QPPIERASAFGNVPLPREAAVDSIDRIYVDAHDRIVQRF-----AMPQTQTAWNVIYTAL 220
Query: 119 YESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRK 178
+LP I G F F E+ + +T DLL+ ADG S++R +
Sbjct: 221 KRALPAGIVHAGDAFERFEQDGERIVAHFASGHAETA-------DLLIGADGGRSAVRAQ 273
Query: 179 YLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYF--DLASGTHSVLYE 236
+PD + Y+GY AWRG++D + ++ G F L G E
Sbjct: 274 LMPDVRPTYAGYVAWRGLVDERGLPEQVLYLLRERFTFQQGDAHLFLTYLVPGADGAT-E 332
Query: 237 LPNKKLNWIWY------------VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPEL 284
+++NW+WY + + + G+ + D ++ A ++ P L
Sbjct: 333 AGKRRVNWVWYRRLASDRLPSLFLARDGTQRDGSLPPGAMRDDNRAELVDAAGRMLAPTL 392
Query: 285 AKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKC 344
A ++ T PF I D +E++ VL+G+AA PH DA L +
Sbjct: 393 AALVDATPAPFAQAIQDL-AVERMAVGRAVLLGDAACLVRPHTAAGVAKAADDAVGLAQA 451
Query: 345 IEKWG 349
+ + G
Sbjct: 452 LREVG 456
>UniRef100_Q7WEZ8 Putative hydroxylase [Bordetella bronchiseptica]
Length = 406
Score = 89.4 bits (220), Expect = 2e-16
Identities = 93/372 (25%), Positives = 164/372 (44%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRDLLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + E+ V A + + I GD+LV ADG S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDI----EQDARQVTATLA---DGTRIQGDILVGADGIHSLVRSRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W ++ +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNEQVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>UniRef100_UPI000031B0F6 UPI000031B0F6 UniRef100 entry
Length = 233
Score = 87.8 bits (216), Expect = 5e-16
Identities = 73/252 (28%), Positives = 121/252 (47%), Gaps = 27/252 (10%)
Query: 1 MVGEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLN-SLSQ 59
M + +A+IVGGS+GG+ +A+ L+ GW V V E+ G GAG+ + L +
Sbjct: 1 MTHASTRARAIIVGGSLGGLFAANLLVRNGWRVDVFERVPEELAGR--GAGIVTHPELFE 58
Query: 60 QIIQSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLY 119
+ + I +T QN S + + +TLT WG ++ VL
Sbjct: 59 ALAAAGIDFDDSIGVKVQSRVTFAQNGAVLSARDLPQTLTA----------WGKMYHVLR 108
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
+LP + G + EK V + G V+ DL++AADG SSIR ++
Sbjct: 109 AALPDVHYHAGGTVAAVQDGPEKALV-----TLADGTVLH--ADLVIAADGFKSSIREQF 161
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAY----PDLGKCLYFDLASGTHSVLY 235
LPD KL+Y+GY AWRG++D + + + ET + + + + P + L + +A +S
Sbjct: 162 LPDVKLQYAGYVAWRGLVDEAAL-SRETREALFEKFAFCLPPREQILGYPVAGQGNST-- 218
Query: 236 ELPNKKLNWIWY 247
++ N++WY
Sbjct: 219 TPGERRYNFVWY 230
>UniRef100_Q7W3M2 Putative hydroxylase [Bordetella parapertussis]
Length = 406
Score = 87.0 bits (214), Expect = 9e-16
Identities = 92/372 (24%), Positives = 163/372 (43%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRDLLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + E+ V A + + I GD+LV AD S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDI----EQDARQVTATLA---DGTRIQGDILVGADSIHSLVRSRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W ++ +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNEQVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>UniRef100_O30447 Hypothetical protein [Bordetella pertussis]
Length = 406
Score = 86.3 bits (212), Expect = 2e-15
Identities = 89/372 (23%), Positives = 159/372 (41%), Gaps = 44/372 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
+ +I G IGG + A AL D +VLE+ P GAG+ L+ ++Q
Sbjct: 2 RVIIAGCGIGGAALAVALEKFKIDHVVLEQA---PRLEEVGAGVQLSPNGVAVLQHL--- 55
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESL-----NFRA----AHWGDLHGVLY 119
+H + + + + + + L R+ + +F A AH DL GVL
Sbjct: 56 ---GVHEALSKVAFEPRELLYRDWQSGQVLMRNPLMPTIKEHFGAPYYHAHRADLLGVLT 112
Query: 120 ESLPPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY 179
E L P G + + + + GD+LV ADG S +R ++
Sbjct: 113 ERLDPAKLRLGSRIVDIDQD-------ARQVTATLADGTRVQGDILVGADGIHSLVRGRF 165
Query: 180 LPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPN 239
+ + SG AWRG++D + D+ + L +V+Y +
Sbjct: 166 FQADQPQASGCIAWRGIVDADAARHL-----------DISPSAHLWLGPERSAVIYYVSG 214
Query: 240 -KKLNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF 298
+K+NWI ++P + S TT V D + + + W + +++ T +PF+
Sbjct: 215 GRKINWICIGSRPGDRKESWSATTTV--DEVLREYAG----WNELVTGLIRLTDKPFVTA 268
Query: 299 IYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALE 358
+YD PL+ + L+G++AH P++ + ++ DA VL + +++ G + + ALE
Sbjct: 269 LYDRAPLDSWINGRIALLGDSAHAMLPYHAQGAVQSMEDAWVLARTLQQSGGD-IPPALE 327
Query: 359 EYQLTRLPVTSK 370
YQ R T++
Sbjct: 328 RYQSLRKDRTAR 339
>UniRef100_Q739B2 FAD binding-monooxygenase family protein [Bacillus cereus]
Length = 377
Score = 81.3 bits (199), Expect = 5e-14
Identities = 88/376 (23%), Positives = 153/376 (40%), Gaps = 40/376 (10%)
Query: 11 VIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-ISKP 69
+I+GG I G+ +A +L G +V V +K T P G G+ + + Q ++ + IS+
Sbjct: 5 IIIGGGIAGLCAAISLQKIGIEVKVYDKNTEPTVA---GTGIIIAPNAMQALEPYGISEK 61
Query: 70 PQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFLW 129
+ N + NLV++ N+ + + H DLH +L L W
Sbjct: 62 IKKFGNESDGF----NLVSEKGTVFNKLIIPSCYPKMYSIHRKDLHQLLLSELQEDTVEW 117
Query: 130 GHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYSG 189
+ NE+ + + V + G E G++L++ADG S +R++ RY+G
Sbjct: 118 RKECVKIE-RNEEDALKI---VFQDGS--EAFGNILISADGIHSVVRKQVTQRDNYRYAG 171
Query: 190 YCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHS--VLYELPNKKLNWIWY 247
Y WRG+ + + + F GT+ + LPN ++ W
Sbjct: 172 YTCWRGITPTNNLSLTND----------------FIETWGTNGRFGIVPLPNNEVYWYAL 215
Query: 248 VNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIYDSDPLEK 307
+N + K + TT+ + + H IP + + I D P+ +
Sbjct: 216 INAKARDPKYKAYTTEDLYNHFKSYHNP-----IPSILHNASDV-HMIHRDIVDIMPMNQ 269
Query: 308 IFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRLPV 367
F +V +G+AAH TP+ + I DA +L +CI+ A EY+ R
Sbjct: 270 FFEKRIVFIGDAAHALTPNLGQGACQAIEDAIILAECIK--NNAHYRQAFIEYEQKRRDR 327
Query: 368 TSKQVLHARRLGRIKQ 383
K A +G++ Q
Sbjct: 328 IEKISNTAWTVGKMAQ 343
>UniRef100_Q8CNB7 Monooxygenase [Staphylococcus epidermidis]
Length = 374
Score = 80.5 bits (197), Expect = 9e-14
Identities = 97/378 (25%), Positives = 159/378 (41%), Gaps = 43/378 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSWISK 68
K IVG IGG+++A L G V V EK TS S G G+G N L +++ ++K
Sbjct: 2 KIAIVGAGIGGLTAAALLEEQGHQVKVFEKNTSINELS-AGIGIGDNVL-KKLGHHDLAK 59
Query: 69 PPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESLPPQIFL 128
+ N LT N+ + + + SLN + L E + +
Sbjct: 60 G---IKNAGQNLTA-MNIYDEQGTPLMSAKLKSHSLNVALSRQ-----TLIEIIQSYVEE 110
Query: 129 WGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPDFKLRYS 188
I F V + K + T + E DL + ADG S +R K+RY+
Sbjct: 111 -SSIHTGFKVTKIE-QTSCKVTLHFTKQESESF-DLCIGADGLHSVVRESVGARTKIRYN 167
Query: 189 GYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKLNWIWYV 248
GY +RG+++ + + +G + L N++ W V
Sbjct: 168 GYTCFRGMVEDVQFNDQHVANEYWGVKGRVG--------------IVPLINQRAYWFITV 213
Query: 249 NQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIP-ELAKVMKETKEP--FLNFIYDSDPL 305
+ E + K S K H +A P E+ KV++ E L+ IYD PL
Sbjct: 214 HAKEGDPKYQS---------FGKPHLQAYFNHFPDEVRKVLERQSETGILLHDIYDLKPL 264
Query: 306 EKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEKWGTEMLESALEEYQLTRL 365
+ + +L+G+AAH TTP+ + + + DA VL C+EK+ A+E Y R+
Sbjct: 265 KTFVYGRTILMGDAAHATTPNMGQGASQAMEDAIVLVNCLEKYD---FNKAIERYDKLRV 321
Query: 366 PVTSKQVLHARRLGRIKQ 383
T+K + ++++G++ Q
Sbjct: 322 KHTAKVIRRSKKIGKMAQ 339
>UniRef100_UPI00002F808F UPI00002F808F UniRef100 entry
Length = 299
Score = 77.8 bits (190), Expect = 6e-13
Identities = 65/222 (29%), Positives = 99/222 (44%), Gaps = 22/222 (9%)
Query: 159 EIVGDLLVAADGCLSSIRRKYLPDFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDL 218
EI GD+L+ ADG S IR++ +L YSG AWRG+ D S+I P +
Sbjct: 88 EISGDILIGADGVFSKIRQQIFSRTELNYSGNFAWRGLADASKIN-----------CPLI 136
Query: 219 GKCLYFDLASGTHSVLYELPNKKL-NWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAE 277
K + H+V L N L N++ V +P KGTS ++++ E
Sbjct: 137 LKNNLIWIGPKKHAVTTILKNGSLINFVGIVEKPADNKKGTSSDNQISA-------LEDF 189
Query: 278 KIWIPELAKVMKETKEPFLNFIYDSDPLEKIFWDNVVLVGEAAHPTTPHYLRSTNMTILD 337
K W EL ++ I+ + P +K V+L+G+AAHP P + + D
Sbjct: 190 KGWHDELIAILNSANSIHRWPIFSARPFKKWSDGKVILIGDAAHPMVPSMAQGAVQALED 249
Query: 338 AAVLGKCIEKWGTEMLESALEEYQLTRLPVTSK-QVLHARRL 378
AA+L + + + +L + RL TS+ Q L R L
Sbjct: 250 AAILSNLLA--NSSDVTRSLSTFHKARLSRTSRIQSLSKRNL 289
>UniRef100_Q9ZSN7 CTF2B [Arabidopsis thaliana]
Length = 428
Score = 77.0 bits (188), Expect = 9e-13
Identities = 93/396 (23%), Positives = 160/396 (39%), Gaps = 50/396 (12%)
Query: 3 GEGRKPKAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQII 62
G R+ K VIVGG IGG+++A AL G +VLE+ S TG GA L L+ +++
Sbjct: 38 GVDREEKVVIVGGGIGGLATAVALHRLGIRSVVLEQAESLRTG---GASLTLSKNGWRVL 94
Query: 63 QSWISKPPQFLHNTTHPLTIDQNLVTDSEKKVNRTLTRDESLNFRAAHWGDLHGVLYESL 122
+ IS PQ + E + + D+S RA L L L
Sbjct: 95 DA-ISIGPQLRKQFLEMQGVVLKKEDGRELRSFKFKDNDQSQEVRAVERRVLLETLASQL 153
Query: 123 PPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKYLPD 182
PPQ + S + G +++ K + + ++++ DG S + ++
Sbjct: 154 PPQTIRFSSKLESIQSNDANGDTLLQLK-----DGTRFLANIVIGCDGIRSKV-ATWMGF 207
Query: 183 FKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKKL 242
+ +Y GYCA+RG+ F P K Y G + + K+
Sbjct: 208 SEPKYVGYCAFRGLGFF------------PNGQPFQQKVNYI-FGRGLRAGYVPVSATKV 254
Query: 243 NWIWYVNQPE--PEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNFIY 300
W N P P++ ++ K +++ W +L ++ T + ++
Sbjct: 255 YWFITFNSPSLGPQMMDPAILRKEAKELV--------STWPDDLQNLIDLTPDEAIS--- 303
Query: 301 DSDPL-EKIFW---------DNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKCIEK--- 347
PL ++ W VVLVG+A HP TP+ + + D+ +L + +
Sbjct: 304 -RTPLADRWLWPGIAPSASKGRVVLVGDAWHPMTPNLGQGACCALEDSVLLANKLARAIN 362
Query: 348 WGTEMLESALEEYQLTRLPVTSKQVLHARRLGRIKQ 383
GTE +E A+E Y+ R + + A +G++ Q
Sbjct: 363 GGTESVERAMESYRSERWSQVFRLTVLANLVGKLLQ 398
>UniRef100_UPI00002FAF2D UPI00002FAF2D UniRef100 entry
Length = 897
Score = 76.3 bits (186), Expect = 2e-12
Identities = 97/389 (24%), Positives = 170/389 (42%), Gaps = 44/389 (11%)
Query: 9 KAVIVGGSIGGISSAHALILAGWDVLVLEKTTSPPTGSPTGAGLGLNSLSQQIIQSW-IS 67
+ ++VGG IGG ++A AL G+DV VLE S G GL +Q IQ+ +
Sbjct: 403 RIIVVGGGIGGAAAAVALQRKGFDVTVLEADES---FDARAQGYGLTVQAQDAIQAMGVD 459
Query: 68 KPPQFLHNTTHPLTIDQNLVTD--SEKKVNRTLTRDESLN---FRAAHWGDLHGVLYESL 122
+T+H ++ + E +++ R E+ N F L + +++
Sbjct: 460 ISQDDAPSTSHYTFSEKGEIIGFFGEAFGSKSKDRRETQNSGRFIHIPRQVLRSRIIQAV 519
Query: 123 PPQIFLWGHIFLSFHVANEKGPVIVKAKVVKTGEVIEIVGDLLVAADGCLSSIRRKY-LP 181
P W SF N+ VK V + ++ G LLV +DG S++RR+ LP
Sbjct: 520 QPGTIQWSSKLKSFACEND-----VKGVSVTLMDGTKLQGALLVGSDGIFSTVRRQLELP 574
Query: 182 DFKLRYSGYCAWRGVLDFSEIENSETIKGIKKAYPDLGKCLYFDLASGTHSVLYELPNKK 241
+L Y G C G++D E I + L + F+ G + LY +P
Sbjct: 575 GDRLNYVGLCVVLGIVD-------EKIMRV-----HLAERRIFETVDGV-TRLYAMPFTT 621
Query: 242 LNWIWYVNQPEPEVKGTSVTTKVTSDMIQKMHQEAEKIWIPELAKVMKETKEPFLNF--I 299
+ +W ++ P E + + TK + + +++ + K W + K++ +T ++ +
Sbjct: 622 SSTMWQLSFPCSE-ETARMYTKDAASLKKEITRRCAK-WHDPIPKMLMKTPLDCMSGYPV 679
Query: 300 YDSDPL-------EKIFWDNVVLVGEAAHPTTPHYLRSTNMTILDAAVLGKC----IEKW 348
YD + L E V L+G+AAHP TP + N + DA +L + + K
Sbjct: 680 YDRELLEPGILRPENEVHRRVTLIGDAAHPMTPFKAQGANQAMSDAVLLAESLIEGVHKH 739
Query: 349 G-TEMLESALEEYQLTRLPVTSKQVLHAR 376
G + + AL ++ L +S+ V+ +R
Sbjct: 740 GPVDGFDFALPLFEQKMLMRSSRVVIGSR 768
Database: uniref100
Posted date: Jan 5, 2005 1:24 AM
Number of letters in database: 848,049,833
Number of sequences in database: 2,790,947
Lambda K H
0.318 0.136 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 747,241,802
Number of Sequences: 2790947
Number of extensions: 32347073
Number of successful extensions: 97462
Number of sequences better than 10.0: 515
Number of HSP's better than 10.0 without gapping: 230
Number of HSP's successfully gapped in prelim test: 285
Number of HSP's that attempted gapping in prelim test: 96762
Number of HSP's gapped (non-prelim): 709
length of query: 428
length of database: 848,049,833
effective HSP length: 130
effective length of query: 298
effective length of database: 485,226,723
effective search space: 144597563454
effective search space used: 144597563454
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 76 (33.9 bits)
Medicago: description of AC144617.5


